Haoshen Li
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
PropSAM: A Propagation-Based Model for Segmenting Any 3D Objects in Multi-Modal Medical Images
Aug 25, 2024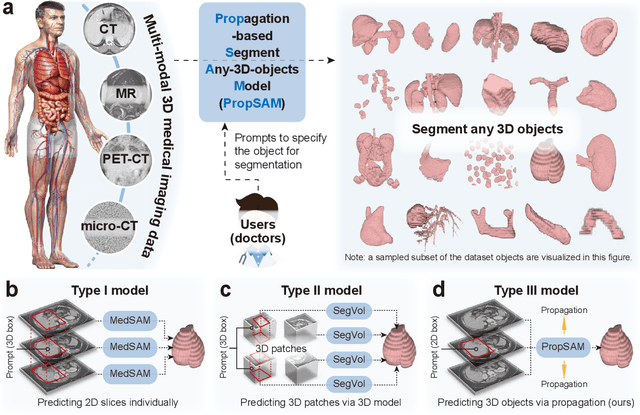
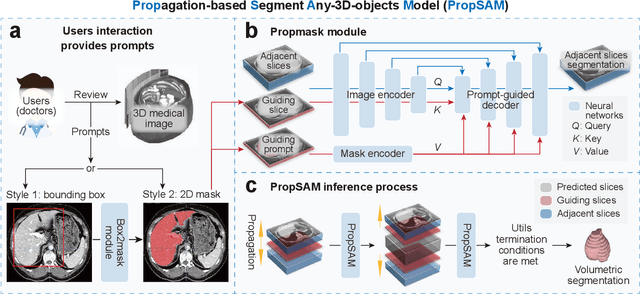
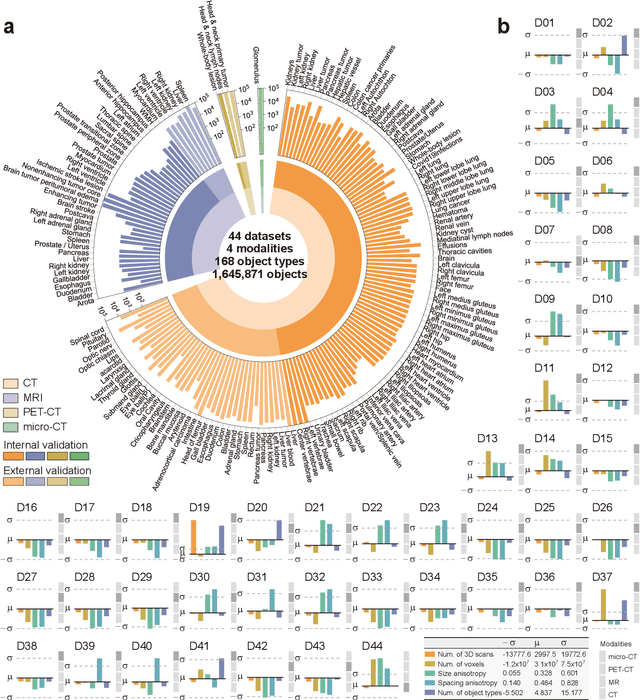
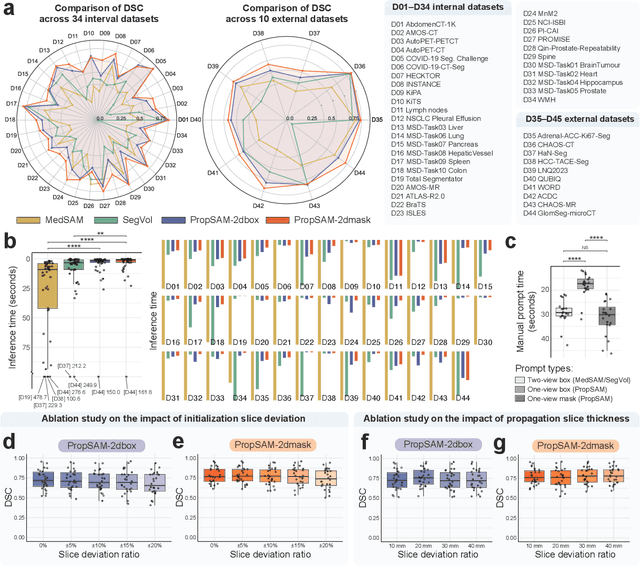
Abstract:Volumetric segmentation is crucial for medical imaging but is often constrained by labor-intensive manual annotations and the need for scenario-specific model training. Furthermore, existing general segmentation models are inefficient due to their design and inferential approaches. Addressing this clinical demand, we introduce PropSAM, a propagation-based segmentation model that optimizes the use of 3D medical structure information. PropSAM integrates a CNN-based UNet for intra-slice processing with a Transformer-based module for inter-slice propagation, focusing on structural and semantic continuities to enhance segmentation across various modalities. Distinctively, PropSAM operates on a one-view prompt, such as a 2D bounding box or sketch mask, unlike conventional models that require two-view prompts. It has demonstrated superior performance, significantly improving the Dice Similarity Coefficient (DSC) across 44 medical datasets and various imaging modalities, outperforming models like MedSAM and SegVol with an average DSC improvement of 18.1%. PropSAM also maintains stable predictions despite prompt deviations and varying propagation configurations, confirmed by one-way ANOVA tests with P>0.5985 and P>0.6131, respectively. Moreover, PropSAM's efficient architecture enables faster inference speeds (Wilcoxon rank-sum test, P<0.001) and reduces user interaction time by 37.8% compared to two-view prompt models. Its ability to handle irregular and complex objects with robust performance further demonstrates its potential in clinical settings, facilitating more automated and reliable medical imaging analyses with minimal retraining.
Effective Lymph Nodes Detection in CT Scans Using Location Debiased Query Selection and Contrastive Query Representation in Transformer
Apr 04, 2024Abstract:Lymph node (LN) assessment is a critical, indispensable yet very challenging task in the routine clinical workflow of radiology and oncology. Accurate LN analysis is essential for cancer diagnosis, staging, and treatment planning. Finding scatteredly distributed, low-contrast clinically relevant LNs in 3D CT is difficult even for experienced physicians under high inter-observer variations. Previous automatic LN detection works typically yield limited recall and high false positives (FPs) due to adjacent anatomies with similar image intensities, shapes, or textures (vessels, muscles, esophagus, etc). In this work, we propose a new LN DEtection TRansformer, named LN-DETR, to achieve more accurate performance. By enhancing the 2D backbone with a multi-scale 2.5D feature fusion to incorporate 3D context explicitly, more importantly, we make two main contributions to improve the representation quality of LN queries. 1) Considering that LN boundaries are often unclear, an IoU prediction head and a location debiased query selection are proposed to select LN queries of higher localization accuracy as the decoder query's initialization. 2) To reduce FPs, query contrastive learning is employed to explicitly reinforce LN queries towards their best-matched ground-truth queries over unmatched query predictions. Trained and tested on 3D CT scans of 1067 patients (with 10,000+ labeled LNs) via combining seven LN datasets from different body parts (neck, chest, and abdomen) and pathologies/cancers, our method significantly improves the performance of previous leading methods by > 4-5% average recall at the same FP rates in both internal and external testing. We further evaluate on the universal lesion detection task using NIH DeepLesion benchmark, and our method achieves the top performance of 88.46% averaged recall across 0.5 to 4 FPs per image, compared with other leading reported results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge