Heng Guo
Near-Light Color Photometric Stereo for mono-Chromaticity non-lambertian surface
Jan 19, 2026Abstract:Color photometric stereo enables single-shot surface reconstruction, extending conventional photometric stereo that requires multiple images of a static scene under varying illumination to dynamic scenarios. However, most existing approaches assume ideal distant lighting and Lambertian reflectance, leaving more practical near-light conditions and non-Lambertian surfaces underexplored. To overcome this limitation, we propose a framework that leverages neural implicit representations for depth and BRDF modeling under the assumption of mono-chromaticity (uniform chromaticity and homogeneous material), which alleviates the inherent ill-posedness of color photometric stereo and allows for detailed surface recovery from just one image. Furthermore, we design a compact optical tactile sensor to validate our approach. Experiments on both synthetic and real-world datasets demonstrate that our method achieves accurate and robust surface reconstruction.
Measurement-Constrained Sampling for Text-Prompted Blind Face Restoration
Nov 18, 2025



Abstract:Blind face restoration (BFR) may correspond to multiple plausible high-quality (HQ) reconstructions under extremely low-quality (LQ) inputs. However, existing methods typically produce deterministic results, struggling to capture this one-to-many nature. In this paper, we propose a Measurement-Constrained Sampling (MCS) approach that enables diverse LQ face reconstructions conditioned on different textual prompts. Specifically, we formulate BFR as a measurement-constrained generative task by constructing an inverse problem through controlled degradations of coarse restorations, which allows posterior-guided sampling within text-to-image diffusion. Measurement constraints include both Forward Measurement, which ensures results align with input structures, and Reverse Measurement, which produces projection spaces, ensuring that the solution can align with various prompts. Experiments show that our MCS can generate prompt-aligned results and outperforms existing BFR methods. Codes will be released after acceptance.
Seeing Through the Rain: Resolving High-Frequency Conflicts in Deraining and Super-Resolution via Diffusion Guidance
Nov 16, 2025Abstract:Clean images are crucial for visual tasks such as small object detection, especially at high resolutions. However, real-world images are often degraded by adverse weather, and weather restoration methods may sacrifice high-frequency details critical for analyzing small objects. A natural solution is to apply super-resolution (SR) after weather removal to recover both clarity and fine structures. However, simply cascading restoration and SR struggle to bridge their inherent conflict: removal aims to remove high-frequency weather-induced noise, while SR aims to hallucinate high-frequency textures from existing details, leading to inconsistent restoration contents. In this paper, we take deraining as a case study and propose DHGM, a Diffusion-based High-frequency Guided Model for generating clean and high-resolution images. DHGM integrates pre-trained diffusion priors with high-pass filters to simultaneously remove rain artifacts and enhance structural details. Extensive experiments demonstrate that DHGM achieves superior performance over existing methods, with lower costs.
SpecGen: Neural Spectral BRDF Generation via Spectral-Spatial Tri-plane Aggregation
Aug 24, 2025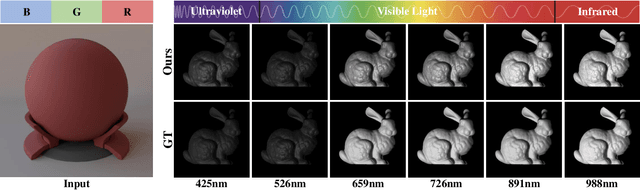
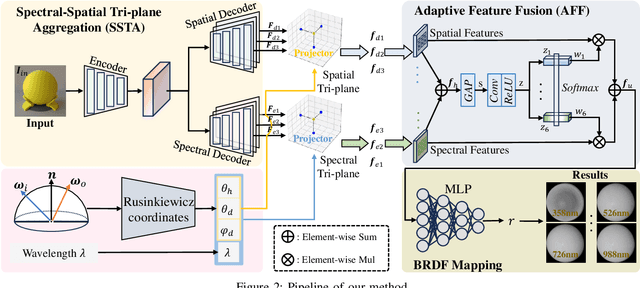
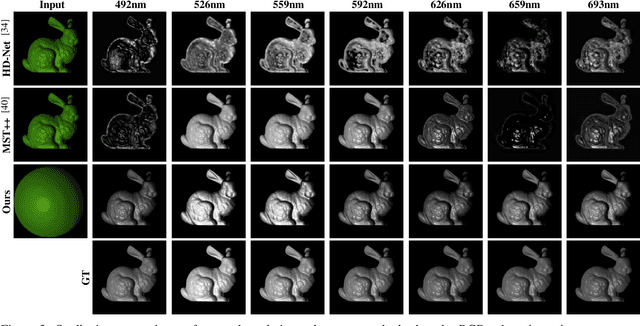
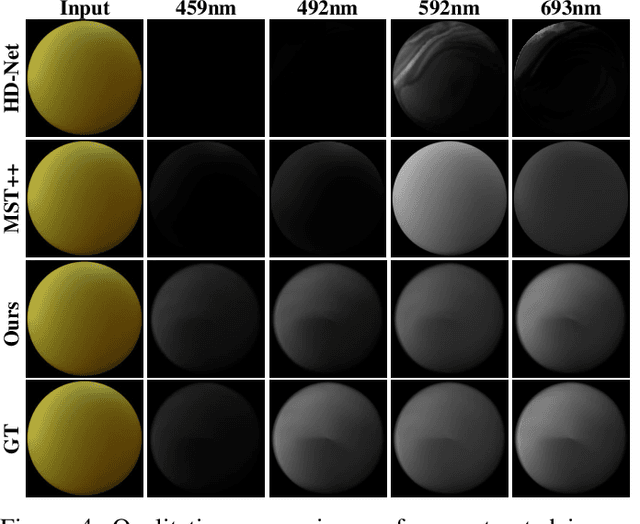
Abstract:Synthesizing spectral images across different wavelengths is essential for photorealistic rendering. Unlike conventional spectral uplifting methods that convert RGB images into spectral ones, we introduce SpecGen, a novel method that generates spectral bidirectional reflectance distribution functions (BRDFs) from a single RGB image of a sphere. This enables spectral image rendering under arbitrary illuminations and shapes covered by the corresponding material. A key challenge in spectral BRDF generation is the scarcity of measured spectral BRDF data. To address this, we propose the Spectral-Spatial Tri-plane Aggregation (SSTA) network, which models reflectance responses across wavelengths and incident-outgoing directions, allowing the training strategy to leverage abundant RGB BRDF data to enhance spectral BRDF generation. Experiments show that our method accurately reconstructs spectral BRDFs from limited spectral data and surpasses state-of-the-art methods in hyperspectral image reconstruction, achieving an improvement of 8 dB in PSNR. Codes and data will be released upon acceptance.
PolarAnything: Diffusion-based Polarimetric Image Synthesis
Jul 24, 2025Abstract:Polarization images facilitate image enhancement and 3D reconstruction tasks, but the limited accessibility of polarization cameras hinders their broader application. This gap drives the need for synthesizing photorealistic polarization images. The existing polarization simulator Mitsuba relies on a parametric polarization image formation model and requires extensive 3D assets covering shape and PBR materials, preventing it from generating large-scale photorealistic images. To address this problem, we propose PolarAnything, capable of synthesizing polarization images from a single RGB input with both photorealism and physical accuracy, eliminating the dependency on 3D asset collections. Drawing inspiration from the zero-shot performance of pretrained diffusion models, we introduce a diffusion-based generative framework with an effective representation strategy that preserves the fidelity of polarization properties. Experiments show that our model generates high-quality polarization images and supports downstream tasks like shape from polarization.
MFogHub: Bridging Multi-Regional and Multi-Satellite Data for Global Marine Fog Detection and Forecasting
May 15, 2025Abstract:Deep learning approaches for marine fog detection and forecasting have outperformed traditional methods, demonstrating significant scientific and practical importance. However, the limited availability of open-source datasets remains a major challenge. Existing datasets, often focused on a single region or satellite, restrict the ability to evaluate model performance across diverse conditions and hinder the exploration of intrinsic marine fog characteristics. To address these limitations, we introduce \textbf{MFogHub}, the first multi-regional and multi-satellite dataset to integrate annotated marine fog observations from 15 coastal fog-prone regions and six geostationary satellites, comprising over 68,000 high-resolution samples. By encompassing diverse regions and satellite perspectives, MFogHub facilitates rigorous evaluation of both detection and forecasting methods under varying conditions. Extensive experiments with 16 baseline models demonstrate that MFogHub can reveal generalization fluctuations due to regional and satellite discrepancy, while also serving as a valuable resource for the development of targeted and scalable fog prediction techniques. Through MFogHub, we aim to advance both the practical monitoring and scientific understanding of marine fog dynamics on a global scale. The dataset and code are at \href{https://github.com/kaka0910/MFogHub}{https://github.com/kaka0910/MFogHub}.
NTIRE 2025 Challenge on Day and Night Raindrop Removal for Dual-Focused Images: Methods and Results
Apr 19, 2025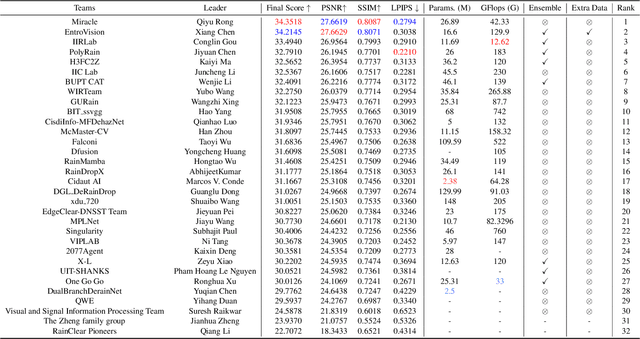
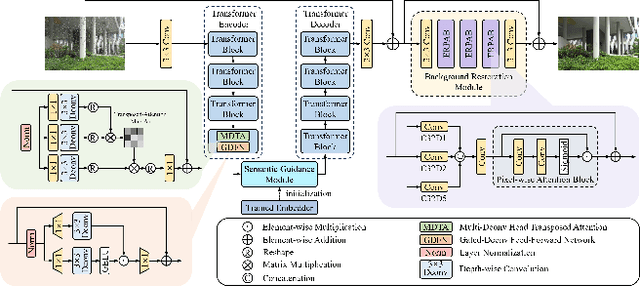
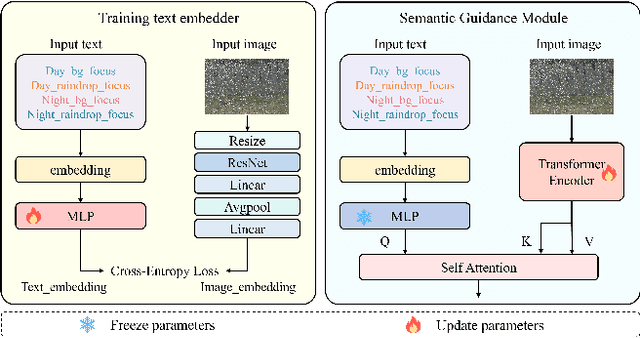
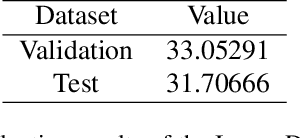
Abstract:This paper reviews the NTIRE 2025 Challenge on Day and Night Raindrop Removal for Dual-Focused Images. This challenge received a wide range of impressive solutions, which are developed and evaluated using our collected real-world Raindrop Clarity dataset. Unlike existing deraining datasets, our Raindrop Clarity dataset is more diverse and challenging in degradation types and contents, which includes day raindrop-focused, day background-focused, night raindrop-focused, and night background-focused degradations. This dataset is divided into three subsets for competition: 14,139 images for training, 240 images for validation, and 731 images for testing. The primary objective of this challenge is to establish a new and powerful benchmark for the task of removing raindrops under varying lighting and focus conditions. There are a total of 361 participants in the competition, and 32 teams submitting valid solutions and fact sheets for the final testing phase. These submissions achieved state-of-the-art (SOTA) performance on the Raindrop Clarity dataset. The project can be found at https://lixinustc.github.io/CVPR-NTIRE2025-RainDrop-Competition.github.io/.
PMNI: Pose-free Multi-view Normal Integration for Reflective and Textureless Surface Reconstruction
Apr 14, 2025

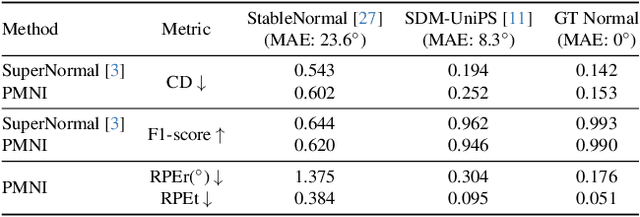

Abstract:Reflective and textureless surfaces remain a challenge in multi-view 3D reconstruction. Both camera pose calibration and shape reconstruction often fail due to insufficient or unreliable cross-view visual features. To address these issues, we present PMNI (Pose-free Multi-view Normal Integration), a neural surface reconstruction method that incorporates rich geometric information by leveraging surface normal maps instead of RGB images. By enforcing geometric constraints from surface normals and multi-view shape consistency within a neural signed distance function (SDF) optimization framework, PMNI simultaneously recovers accurate camera poses and high-fidelity surface geometry. Experimental results on synthetic and real-world datasets show that our method achieves state-of-the-art performance in the reconstruction of reflective surfaces, even without reliable initial camera poses.
PIDSR:ComplementaryPolarizedImageDemosaicingandSuper-Resolution
Apr 10, 2025



Abstract:Polarization cameras can capture multiple polarized images with different polarizer angles in a single shot, bringing convenience to polarization-based downstream tasks. However, their direct outputs are color-polarization filter array (CPFA) raw images, requiring demosaicing to reconstruct full-resolution, full-color polarized images; unfortunately, this necessary step introduces artifacts that make polarization-related parameters such as the degree of polarization (DoP) and angle of polarization (AoP) prone to error. Besides, limited by the hardware design, the resolution of a polarization camera is often much lower than that of a conventional RGB camera. Existing polarized image demosaicing (PID) methods are limited in that they cannot enhance resolution, while polarized image super-resolution (PISR) methods, though designed to obtain high-resolution (HR) polarized images from the demosaicing results, tend to retain or even amplify errors in the DoP and AoP introduced by demosaicing artifacts. In this paper, we propose PIDSR, a joint framework that performs complementary Polarized Image Demosaicing and Super-Resolution, showing the ability to robustly obtain high-quality HR polarized images with more accurate DoP and AoP from a CPFA raw image in a direct manner. Experiments show our PIDSR not only achieves state-of-the-art performance on both synthetic and real data, but also facilitates downstream tasks.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge