Guole Liu
XNet v2: Fewer Limitations, Better Results and Greater Universality
Sep 02, 2024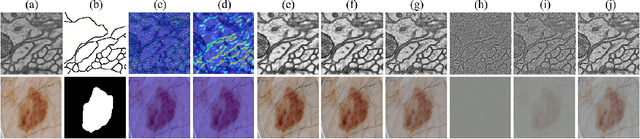
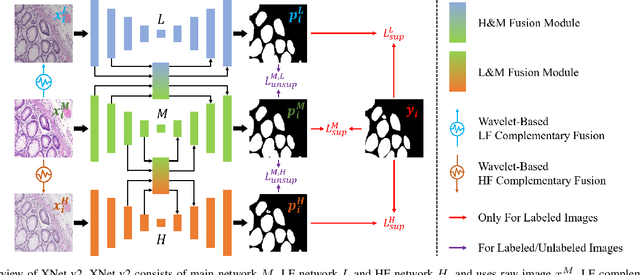
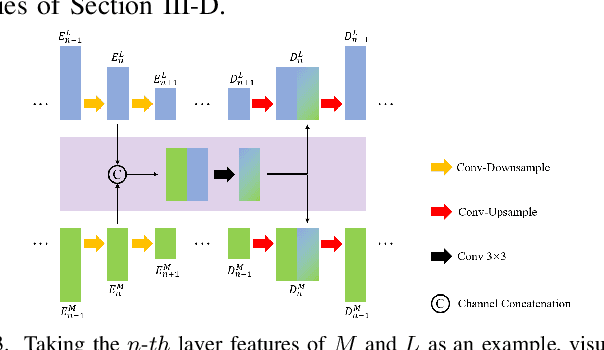
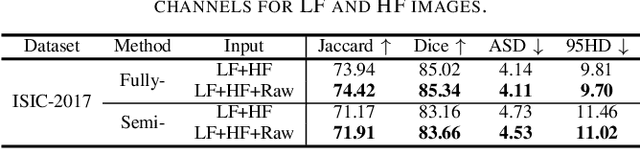
Abstract:XNet introduces a wavelet-based X-shaped unified architecture for fully- and semi-supervised biomedical segmentation. So far, however, XNet still faces the limitations, including performance degradation when images lack high-frequency (HF) information, underutilization of raw images and insufficient fusion. To address these issues, we propose XNet v2, a low- and high-frequency complementary model. XNet v2 performs wavelet-based image-level complementary fusion, using fusion results along with raw images inputs three different sub-networks to construct consistency loss. Furthermore, we introduce a feature-level fusion module to enhance the transfer of low-frequency (LF) information and HF information. XNet v2 achieves state-of-the-art in semi-supervised segmentation while maintaining competitve results in fully-supervised learning. More importantly, XNet v2 excels in scenarios where XNet fails. Compared to XNet, XNet v2 exhibits fewer limitations, better results and greater universality. Extensive experiments on three 2D and two 3D datasets demonstrate the effectiveness of XNet v2. Code is available at https://github.com/Yanfeng-Zhou/XNetv2 .
Representing Topological Self-Similarity Using Fractal Feature Maps for Accurate Segmentation of Tubular Structures
Jul 20, 2024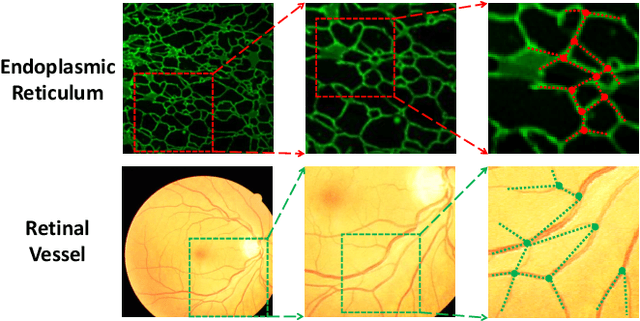
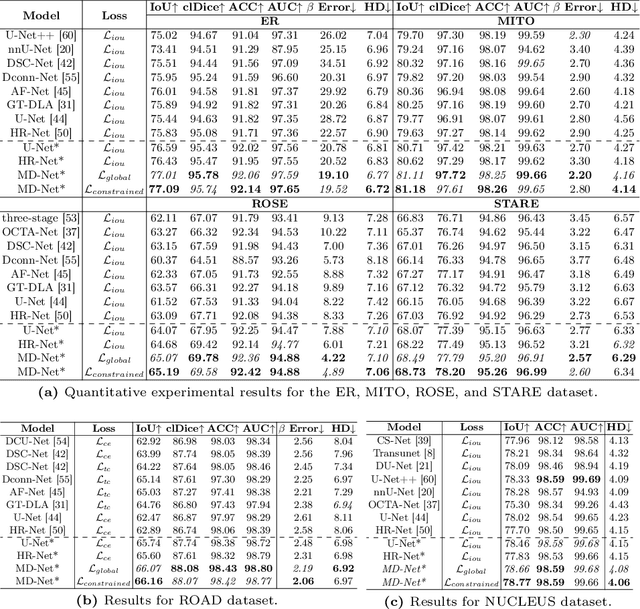
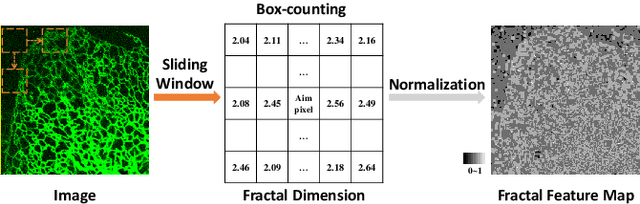
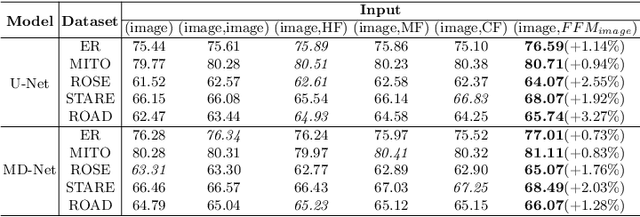
Abstract:Accurate segmentation of long and thin tubular structures is required in a wide variety of areas such as biology, medicine, and remote sensing. The complex topology and geometry of such structures often pose significant technical challenges. A fundamental property of such structures is their topological self-similarity, which can be quantified by fractal features such as fractal dimension (FD). In this study, we incorporate fractal features into a deep learning model by extending FD to the pixel-level using a sliding window technique. The resulting fractal feature maps (FFMs) are then incorporated as additional input to the model and additional weight in the loss function to enhance segmentation performance by utilizing the topological self-similarity. Moreover, we extend the U-Net architecture by incorporating an edge decoder and a skeleton decoder to improve boundary accuracy and skeletal continuity of segmentation, respectively. Extensive experiments on five tubular structure datasets validate the effectiveness and robustness of our approach. Furthermore, the integration of FFMs with other popular segmentation models such as HR-Net also yields performance enhancement, suggesting FFM can be incorporated as a plug-in module with different model architectures. Code and data are openly accessible at https://github.com/cbmi-group/FFM-Multi-Decoder-Network.
ADFA: Attention-augmented Differentiable top-k Feature Adaptation for Unsupervised Medical Anomaly Detection
Aug 29, 2023Abstract:The scarcity of annotated data, particularly for rare diseases, limits the variability of training data and the range of detectable lesions, presenting a significant challenge for supervised anomaly detection in medical imaging. To solve this problem, we propose a novel unsupervised method for medical image anomaly detection: Attention-Augmented Differentiable top-k Feature Adaptation (ADFA). The method utilizes Wide-ResNet50-2 (WR50) network pre-trained on ImageNet to extract initial feature representations. To reduce the channel dimensionality while preserving relevant channel information, we employ an attention-augmented patch descriptor on the extracted features. We then apply differentiable top-k feature adaptation to train the patch descriptor, mapping the extracted feature representations to a new vector space, enabling effective detection of anomalies. Experiments show that ADFA outperforms state-of-the-art (SOTA) methods on multiple challenging medical image datasets, confirming its effectiveness in medical anomaly detection.
Elucidating Meta-Structures of Noisy Labels in Semantic Segmentation by Deep Neural Networks
Apr 30, 2022
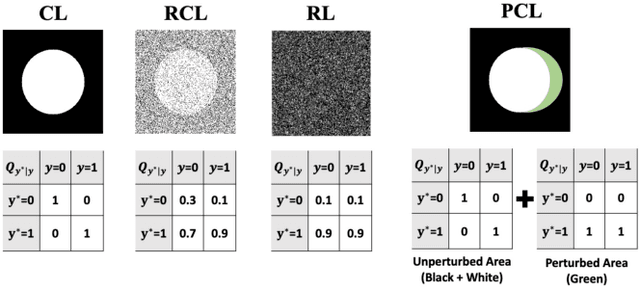
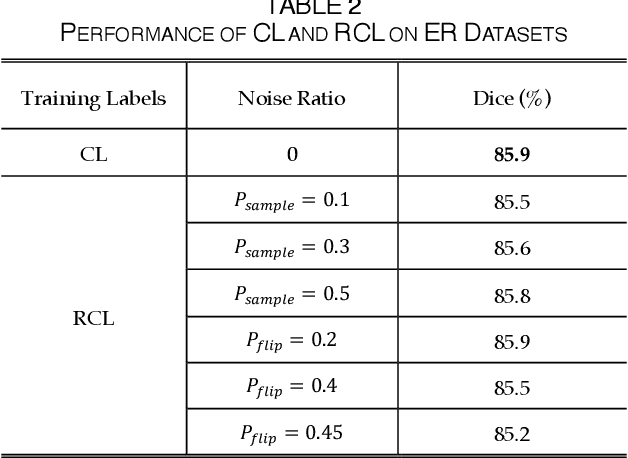
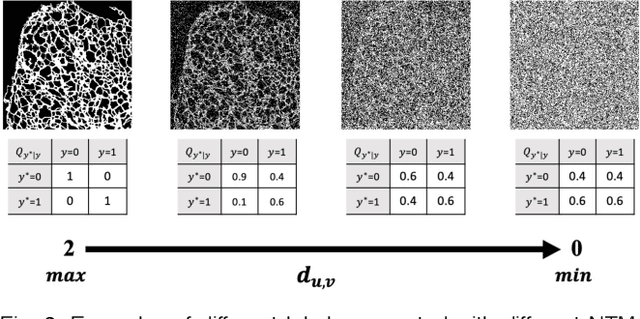
Abstract:The supervised training of deep neural networks (DNNs) by noisy labels has been studied extensively in image classification but much less in image segmentation. So far, our understanding of the learning behavior of DNNs trained by noisy segmentation labels remains limited. In this study, we address this deficiency in both binary segmentation of biological microscopy images and multi-class segmentation of natural images. We classify segmentation labels according to their noise transition matrices (NTM) and compare performance of DNNs trained by different types of labels. When we randomly sample a small fraction (e.g., 10%) or flipping a large fraction (e.g., 90%) of the ground-truth labels to train DNNs, their segmentation performance remains largely the same. This indicates that DNNs learn structures hidden in labels rather than pixel-level labels per se in their supervised training for semantic segmentation. We call these hidden structures "meta-structures". When we use labels with different perturbations to the meta-structures to train DNNs, their performance in feature extraction and segmentation degrades consistently. In contrast, addition of meta-structure information substantially improves performance of an unsupervised model in binary semantic segmentation. We formulate meta-structures mathematically as spatial density distributions and quantify semantic information of different types of labels, which we find to correlate strongly with ranks of their NTM. We show theoretically and experimentally how this formulation explains key observed learning behavior of DNNs.
Deep Neural Networks Learn Meta-Structures to Segment Fluorescence Microscopy Images
Mar 22, 2021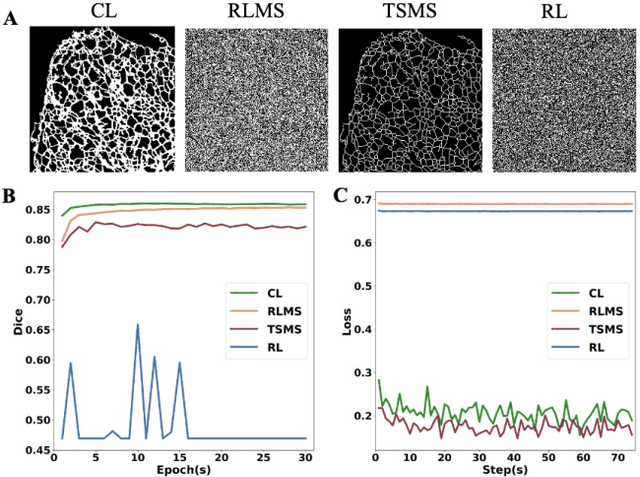
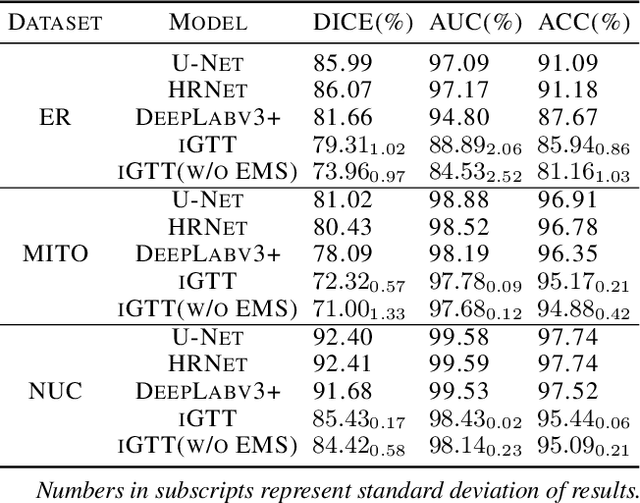
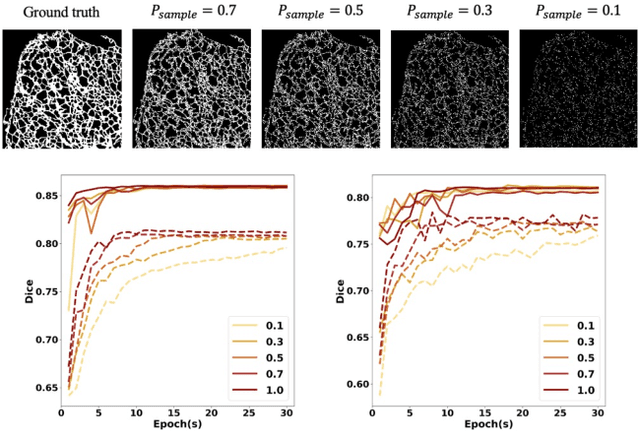
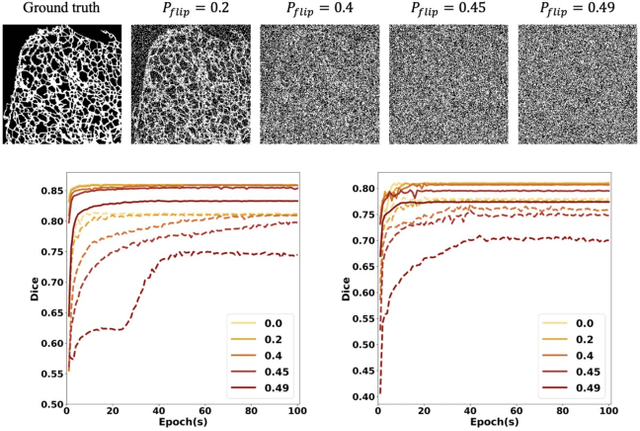
Abstract:Fluorescence microscopy images play the critical role of capturing spatial or spatiotemporal information of biomedical processes in life sciences. Their simple structures and semantics provide unique advantages in elucidating learning behavior of deep neural networks (DNNs). It is generally assumed that accurate image annotation is required to train DNNs for accurate image segmentation. In this study, however, we find that DNNs trained by label images in which nearly half (49%) of the binary pixel labels are randomly flipped provide largely the same segmentation performance. This suggests that DNNs learn high-level structures rather than pixel-level labels per se to segment fluorescence microscopy images. We refer to these structures as meta-structures. In support of the existence of the meta-structures, when DNNs are trained by a series of label images with progressively less meta-structure information, we find progressive degradation in their segmentation performance. Motivated by the learning behavior of DNNs trained by random labels and the characteristics of meta-structures, we propose an unsupervised segmentation model. Experiments show that it achieves remarkably competitive performance in comparison to supervised segmentation models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge