Elucidating Meta-Structures of Noisy Labels in Semantic Segmentation by Deep Neural Networks
Paper and Code
Apr 30, 2022
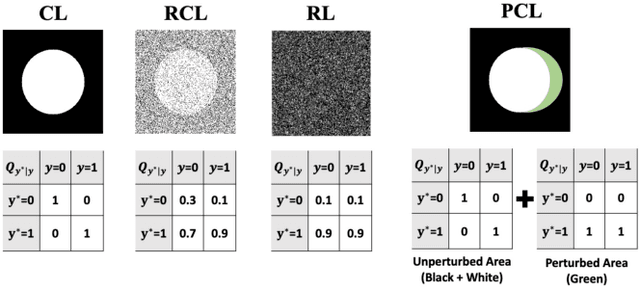
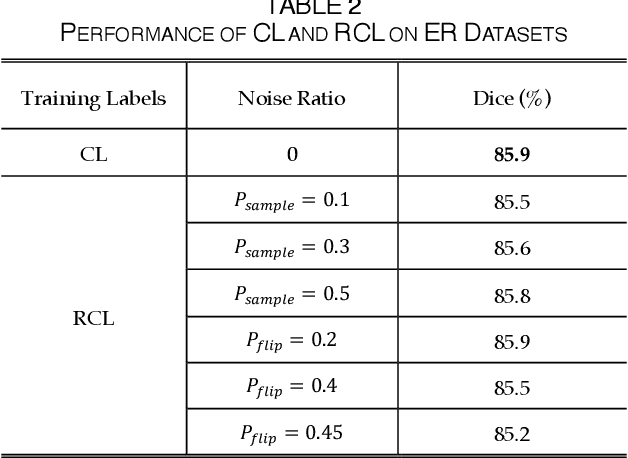
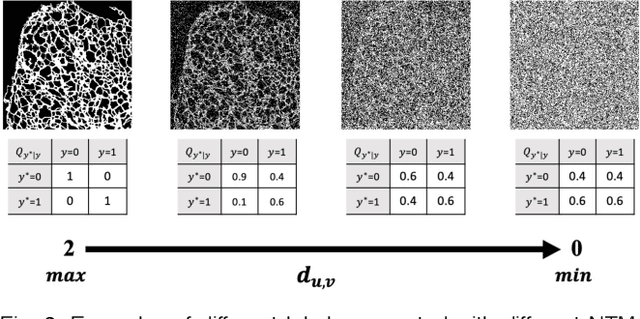
The supervised training of deep neural networks (DNNs) by noisy labels has been studied extensively in image classification but much less in image segmentation. So far, our understanding of the learning behavior of DNNs trained by noisy segmentation labels remains limited. In this study, we address this deficiency in both binary segmentation of biological microscopy images and multi-class segmentation of natural images. We classify segmentation labels according to their noise transition matrices (NTM) and compare performance of DNNs trained by different types of labels. When we randomly sample a small fraction (e.g., 10%) or flipping a large fraction (e.g., 90%) of the ground-truth labels to train DNNs, their segmentation performance remains largely the same. This indicates that DNNs learn structures hidden in labels rather than pixel-level labels per se in their supervised training for semantic segmentation. We call these hidden structures "meta-structures". When we use labels with different perturbations to the meta-structures to train DNNs, their performance in feature extraction and segmentation degrades consistently. In contrast, addition of meta-structure information substantially improves performance of an unsupervised model in binary semantic segmentation. We formulate meta-structures mathematically as spatial density distributions and quantify semantic information of different types of labels, which we find to correlate strongly with ranks of their NTM. We show theoretically and experimentally how this formulation explains key observed learning behavior of DNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge