Yuanhao Guo
Elucidating Meta-Structures of Noisy Labels in Semantic Segmentation by Deep Neural Networks
Apr 30, 2022
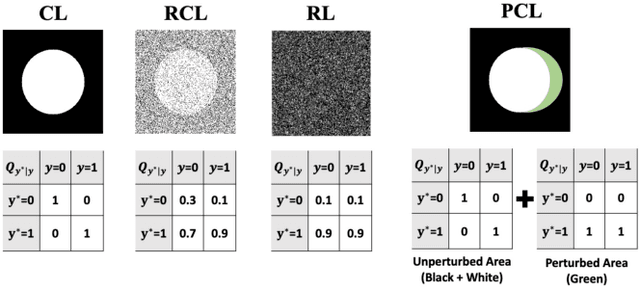
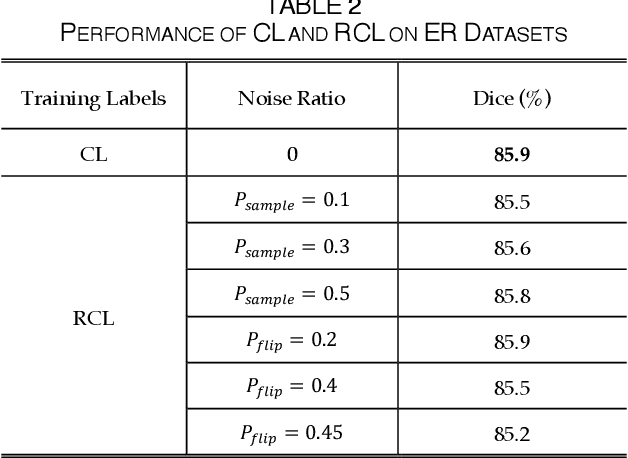
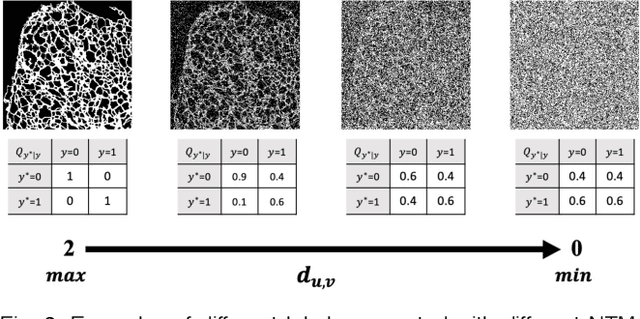
Abstract:The supervised training of deep neural networks (DNNs) by noisy labels has been studied extensively in image classification but much less in image segmentation. So far, our understanding of the learning behavior of DNNs trained by noisy segmentation labels remains limited. In this study, we address this deficiency in both binary segmentation of biological microscopy images and multi-class segmentation of natural images. We classify segmentation labels according to their noise transition matrices (NTM) and compare performance of DNNs trained by different types of labels. When we randomly sample a small fraction (e.g., 10%) or flipping a large fraction (e.g., 90%) of the ground-truth labels to train DNNs, their segmentation performance remains largely the same. This indicates that DNNs learn structures hidden in labels rather than pixel-level labels per se in their supervised training for semantic segmentation. We call these hidden structures "meta-structures". When we use labels with different perturbations to the meta-structures to train DNNs, their performance in feature extraction and segmentation degrades consistently. In contrast, addition of meta-structure information substantially improves performance of an unsupervised model in binary semantic segmentation. We formulate meta-structures mathematically as spatial density distributions and quantify semantic information of different types of labels, which we find to correlate strongly with ranks of their NTM. We show theoretically and experimentally how this formulation explains key observed learning behavior of DNNs.
REFUGE2 Challenge: Treasure for Multi-Domain Learning in Glaucoma Assessment
Feb 24, 2022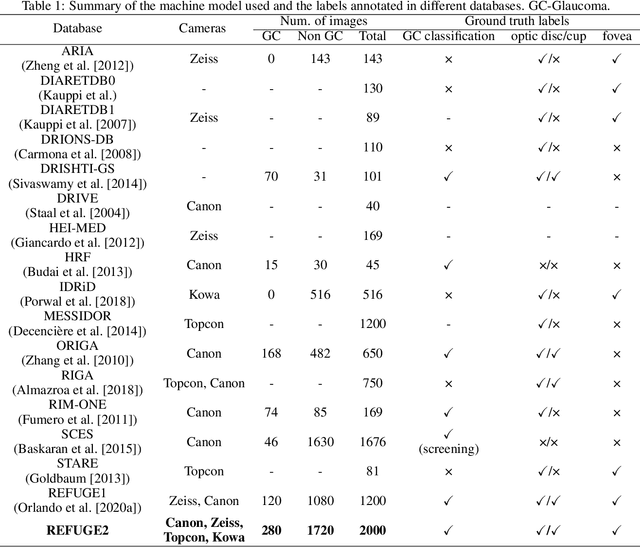
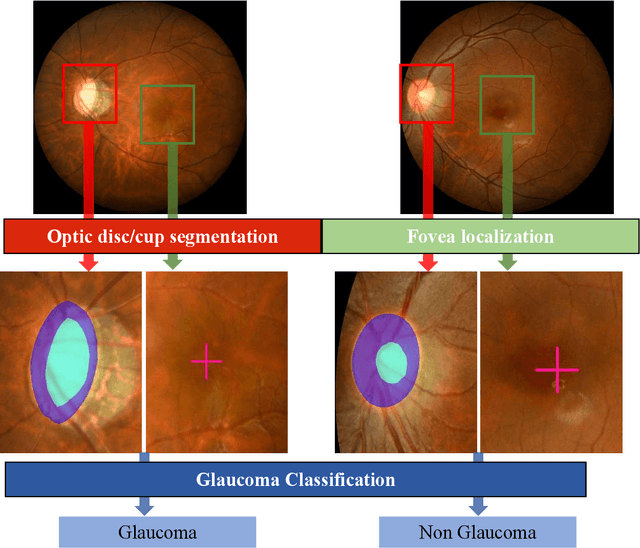
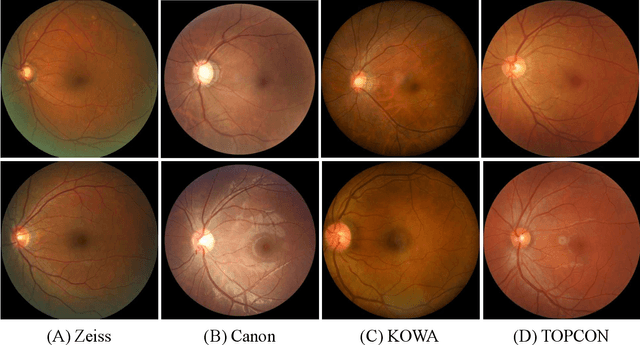
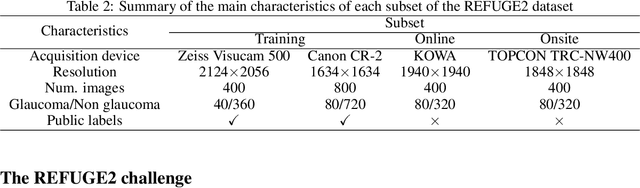
Abstract:Glaucoma is the second leading cause of blindness and is the leading cause of irreversible blindness disease in the world. Early screening for glaucoma in the population is significant. Color fundus photography is the most cost effective imaging modality to screen for ocular diseases. Deep learning network is often used in color fundus image analysis due to its powful feature extraction capability. However, the model training of deep learning method needs a large amount of data, and the distribution of data should be abundant for the robustness of model performance. To promote the research of deep learning in color fundus photography and help researchers further explore the clinical application signification of AI technology, we held a REFUGE2 challenge. This challenge released 2,000 color fundus images of four models, including Zeiss, Canon, Kowa and Topcon, which can validate the stabilization and generalization of algorithms on multi-domain. Moreover, three sub-tasks were designed in the challenge, including glaucoma classification, cup/optic disc segmentation, and macular fovea localization. These sub-tasks technically cover the three main problems of computer vision and clinicly cover the main researchs of glaucoma diagnosis. Over 1,300 international competitors joined the REFUGE2 challenge, 134 teams submitted more than 3,000 valid preliminary results, and 22 teams reached the final. This article summarizes the methods of some of the finalists and analyzes their results. In particular, we observed that the teams using domain adaptation strategies had high and robust performance on the dataset with multi-domain. This indicates that UDA and other multi-domain related researches will be the trend of deep learning field in the future, and our REFUGE2 datasets will play an important role in these researches.
Deep Neural Networks Learn Meta-Structures to Segment Fluorescence Microscopy Images
Mar 22, 2021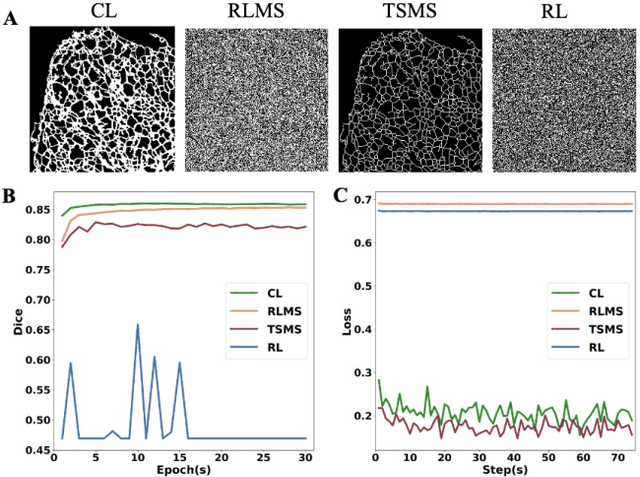
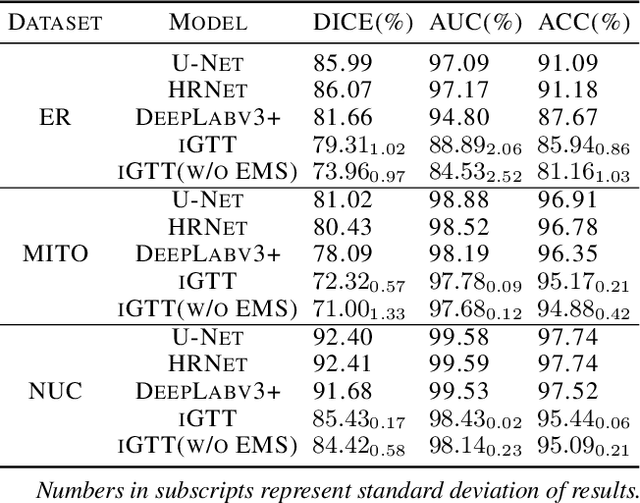
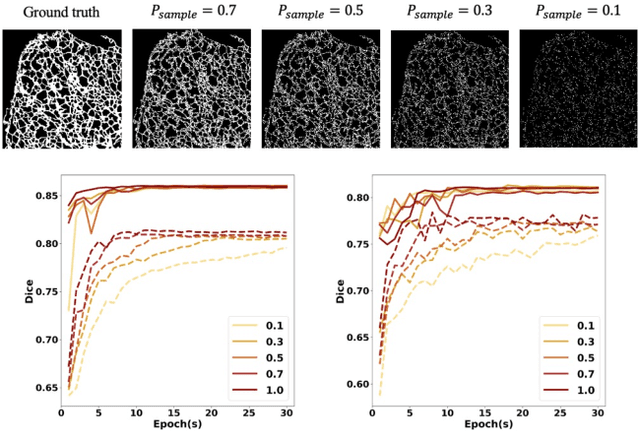
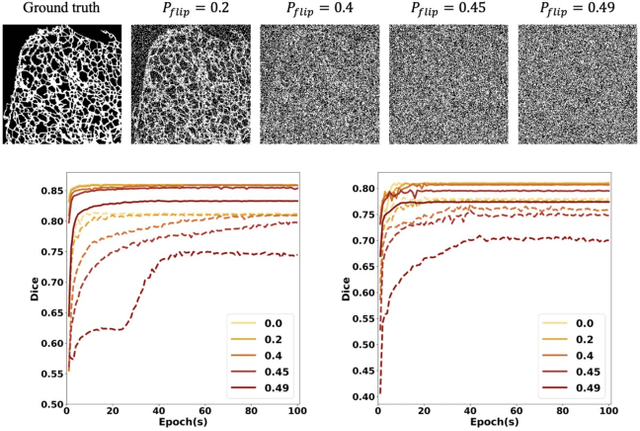
Abstract:Fluorescence microscopy images play the critical role of capturing spatial or spatiotemporal information of biomedical processes in life sciences. Their simple structures and semantics provide unique advantages in elucidating learning behavior of deep neural networks (DNNs). It is generally assumed that accurate image annotation is required to train DNNs for accurate image segmentation. In this study, however, we find that DNNs trained by label images in which nearly half (49%) of the binary pixel labels are randomly flipped provide largely the same segmentation performance. This suggests that DNNs learn high-level structures rather than pixel-level labels per se to segment fluorescence microscopy images. We refer to these structures as meta-structures. In support of the existence of the meta-structures, when DNNs are trained by a series of label images with progressively less meta-structure information, we find progressive degradation in their segmentation performance. Motivated by the learning behavior of DNNs trained by random labels and the characteristics of meta-structures, we propose an unsupervised segmentation model. Experiments show that it achieves remarkably competitive performance in comparison to supervised segmentation models.
Temporal Self-Ensembling Teacher for Semi-Supervised Object Detection
Jul 14, 2020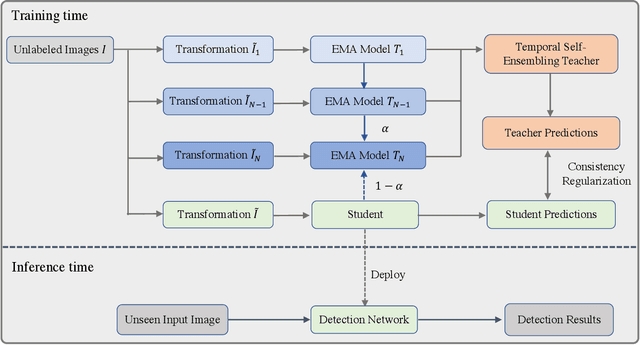
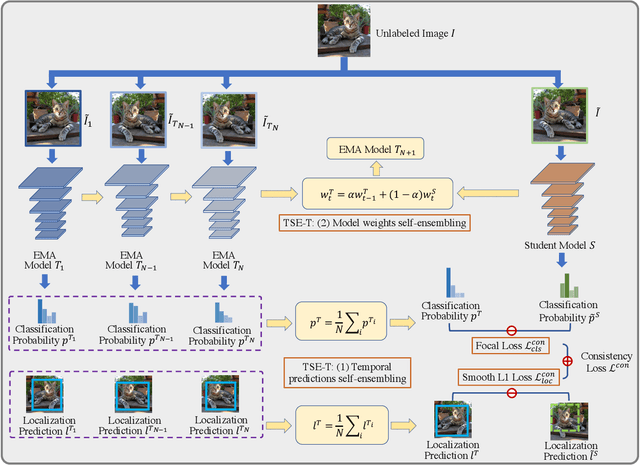
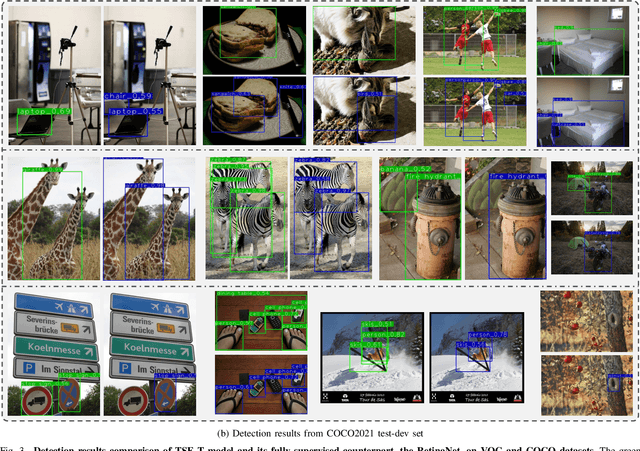

Abstract:This paper focuses on the problem of Semi-Supervised Object Detection (SSOD). In the field of Semi-Supervised Learning (SSL), the Knowledge Distillation (KD) framework which consists of a teacher model and a student model is widely used to make good use of the unlabeled images. Given unlabeled images, the teacher is supposed to yield meaningful targets (e.g. well-posed logits) to regularize the training of the student. However, directly applying the KD framework in SSOD has the following obstacles. (1) Teacher and student predictions may be very close which limits the upper-bound of the student, and (2) the data imbalance dilemma caused by dense prediction from object detection hinders an efficient consistency regularization between the teacher and student. To solve these problems, we propose the Temporal Self-Ensembling Teacher (TSE-T) model on top of the KD framework. Differently from the conventional KD methods, we devise a temporally updated teacher model. First, our teacher model ensembles its temporal predictions for unlabeled images under varying perturbations. Second, our teacher model ensembles its temporal model weights by Exponential Moving Average (EMA) which allows it gradually learn from student. The above self-ensembling strategies collaboratively yield better teacher predictions for unblabeled images. Finally, we use focal loss to formulate the consistency regularization to handle the data imbalance problem. Evaluated on the widely used VOC and COCO benchmarks, our method has achieved 80.73% and 40.52% (mAP) on the VOC2007 test set and the COCO2012 test-dev set respectively, which outperforms the fully-supervised detector by 2.37% and 1.49%. Furthermore, our method sets the new state state of the art in SSOD on VOC benchmark which outperforms the baseline SSOD method by 1.44%. The source code of this work is publicly available at http://github.com/SYangDong/tse-t.
Probabilistic Inference for Camera Calibration in Light Microscopy under Circular Motion
Oct 30, 2019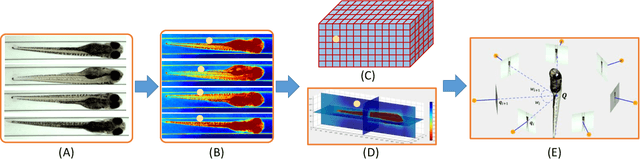
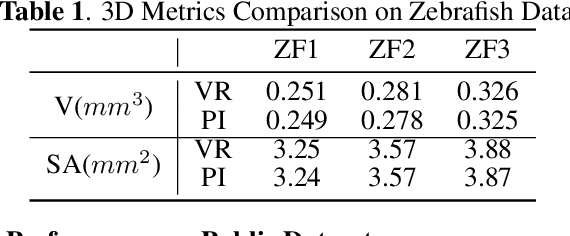
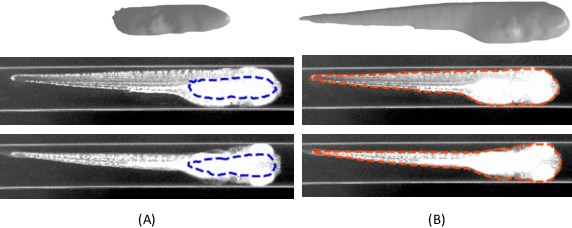
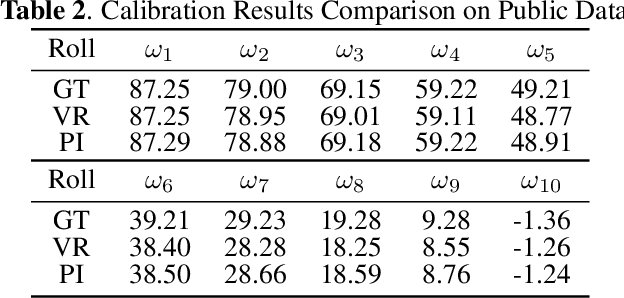
Abstract:Robust and accurate camera calibration is essential for 3D reconstruction in light microscopy under circular motion. Conventional methods require either accurate key point matching or precise segmentation of the axial-view images. Both remain challenging because specimens often exhibit transparency/translucency in a light microscope. To address those issues, we propose a probabilistic inference based method for the camera calibration that does not require sophisticated image pre-processing. Based on 3D projective geometry, our method assigns a probability on each of a range of voxels that cover the whole object. The probability indicates the likelihood of a voxel belonging to the object to be reconstructed. Our method maximizes a joint probability that distinguishes the object from the background. Experimental results show that the proposed method can accurately recover camera configurations in both light microscopy and natural scene imaging. Furthermore, the method can be used to produce high-fidelity 3D reconstructions and accurate 3D measurements.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge