Ziwen Liu
waveOrder: generalist framework for label-agnostic computational microscopy
Dec 13, 2024



Abstract:Correlative computational microscopy is accelerating the mapping of dynamic biological systems by integrating morphological and molecular measurements across spatial scales, from organelles to entire organisms. Visualization, measurement, and prediction of interactions among the components of biological systems can be accelerated by generalist computational imaging frameworks that relax the trade-offs imposed by multiplex dynamic imaging. This work reports a generalist framework for wave optical imaging of the architectural order (waveOrder) among biomolecules for encoding and decoding multiple specimen properties from a minimal set of acquired channels, with or without fluorescent labels. waveOrder expresses material properties in terms of elegant physically motivated basis vectors directly interpretable as phase, absorption, birefringence, diattenuation, and fluorophore density; and it expresses image data in terms of directly measurable Stokes parameters. We report a corresponding multi-channel reconstruction algorithm to recover specimen properties in multiple contrast modes. With this framework, we implement multiple 3D computational microscopy methods, including quantitative phase imaging, quantitative label-free imaging with phase and polarization, and fluorescence deconvolution imaging, across scales ranging from organelles to whole zebrafish. These advances are available via an extensible open-source computational imaging library, waveOrder, and a napari plugin, recOrder.
Contrastive learning of cell state dynamics in response to perturbations
Oct 15, 2024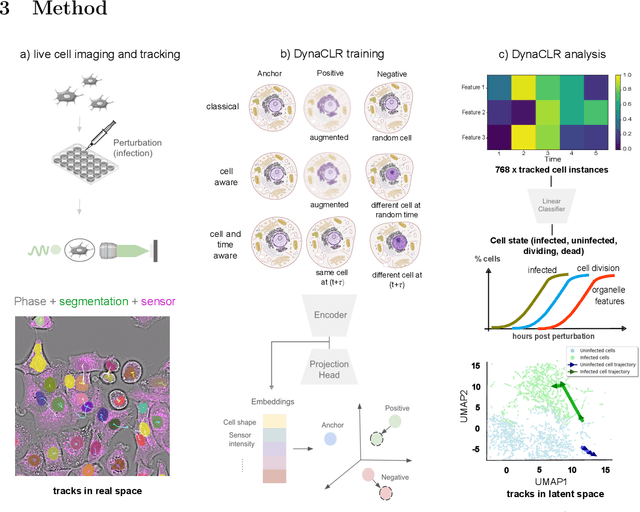
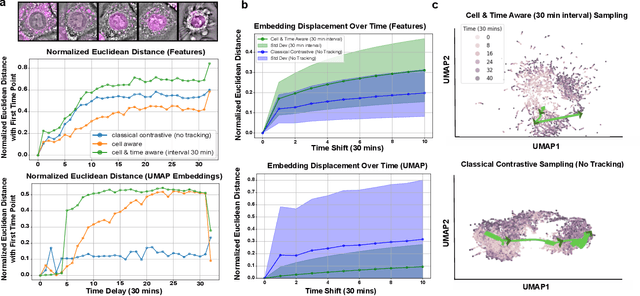
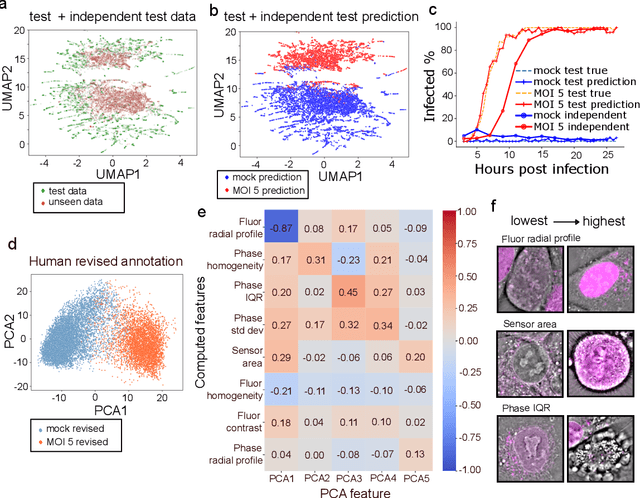
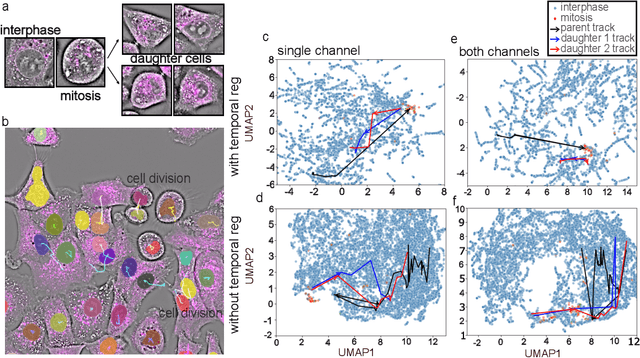
Abstract:We introduce DynaCLR, a self-supervised framework for modeling cell dynamics via contrastive learning of representations of time-lapse datasets. Live cell imaging of cells and organelles is widely used to analyze cellular responses to perturbations. Human annotation of dynamic cell states captured by time-lapse perturbation datasets is laborious and prone to bias. DynaCLR integrates single-cell tracking with time-aware contrastive learning to map images of cells at neighboring time points to neighboring embeddings. Mapping the morphological dynamics of cells to a temporally regularized embedding space makes the annotation, classification, clustering, or interpretation of the cell states more quantitative and efficient. We illustrate the features and applications of DynaCLR with the following experiments: analyzing the kinetics of viral infection in human cells, detecting transient changes in cell morphology due to cell division, and mapping the dynamics of organelles due to viral infection. Models trained with DynaCLR consistently achieve $>95\%$ accuracy for infection state classification, enable the detection of transient cell states and reliably embed unseen experiments. DynaCLR provides a flexible framework for comparative analysis of cell state dynamics due to perturbations, such as infection, gene knockouts, and drugs. We provide PyTorch-based implementations of the model training and inference pipeline (https://github.com/mehta-lab/viscy) and a user interface (https://github.com/czbiohub-sf/napari-iohub) for the visualization and annotation of trajectories of cells in the real space and the embedding space.
XNet v2: Fewer Limitations, Better Results and Greater Universality
Sep 02, 2024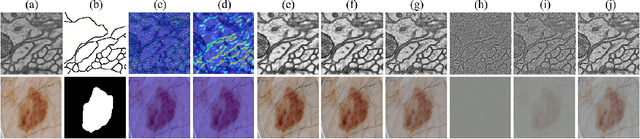
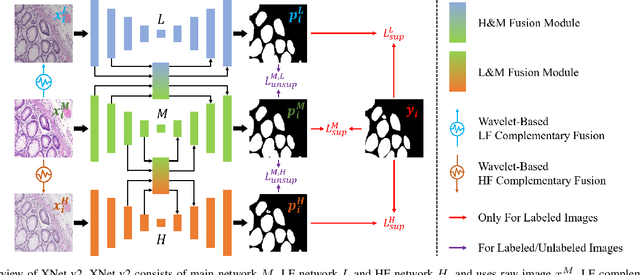
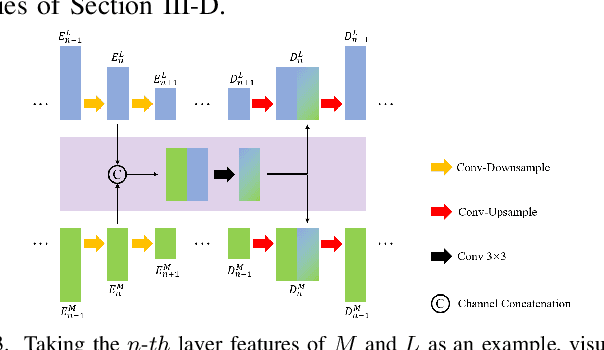
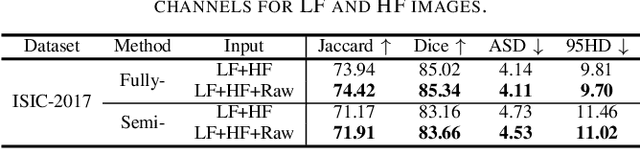
Abstract:XNet introduces a wavelet-based X-shaped unified architecture for fully- and semi-supervised biomedical segmentation. So far, however, XNet still faces the limitations, including performance degradation when images lack high-frequency (HF) information, underutilization of raw images and insufficient fusion. To address these issues, we propose XNet v2, a low- and high-frequency complementary model. XNet v2 performs wavelet-based image-level complementary fusion, using fusion results along with raw images inputs three different sub-networks to construct consistency loss. Furthermore, we introduce a feature-level fusion module to enhance the transfer of low-frequency (LF) information and HF information. XNet v2 achieves state-of-the-art in semi-supervised segmentation while maintaining competitve results in fully-supervised learning. More importantly, XNet v2 excels in scenarios where XNet fails. Compared to XNet, XNet v2 exhibits fewer limitations, better results and greater universality. Extensive experiments on three 2D and two 3D datasets demonstrate the effectiveness of XNet v2. Code is available at https://github.com/Yanfeng-Zhou/XNetv2 .
Blaze3DM: Marry Triplane Representation with Diffusion for 3D Medical Inverse Problem Solving
May 24, 2024



Abstract:Solving 3D medical inverse problems such as image restoration and reconstruction is crucial in modern medical field. However, the curse of dimensionality in 3D medical data leads mainstream volume-wise methods to suffer from high resource consumption and challenges models to successfully capture the natural distribution, resulting in inevitable volume inconsistency and artifacts. Some recent works attempt to simplify generation in the latent space but lack the capability to efficiently model intricate image details. To address these limitations, we present Blaze3DM, a novel approach that enables fast and high-fidelity generation by integrating compact triplane neural field and powerful diffusion model. In technique, Blaze3DM begins by optimizing data-dependent triplane embeddings and a shared decoder simultaneously, reconstructing each triplane back to the corresponding 3D volume. To further enhance 3D consistency, we introduce a lightweight 3D aware module to model the correlation of three vertical planes. Then, diffusion model is trained on latent triplane embeddings and achieves both unconditional and conditional triplane generation, which is finally decoded to arbitrary size volume. Extensive experiments on zero-shot 3D medical inverse problem solving, including sparse-view CT, limited-angle CT, compressed-sensing MRI, and MRI isotropic super-resolution, demonstrate that Blaze3DM not only achieves state-of-the-art performance but also markedly improves computational efficiency over existing methods (22~40x faster than previous work).
Learning Dynamic Tetrahedra for High-Quality Talking Head Synthesis
Feb 27, 2024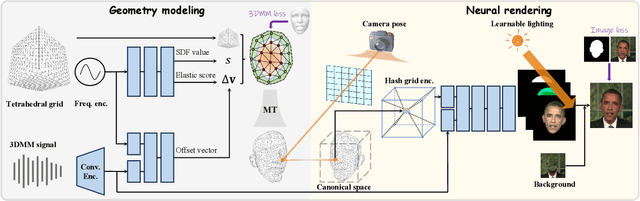
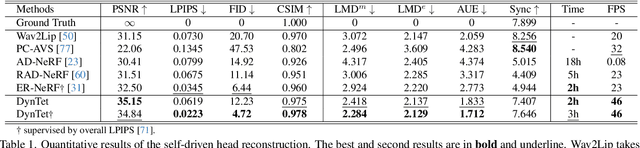

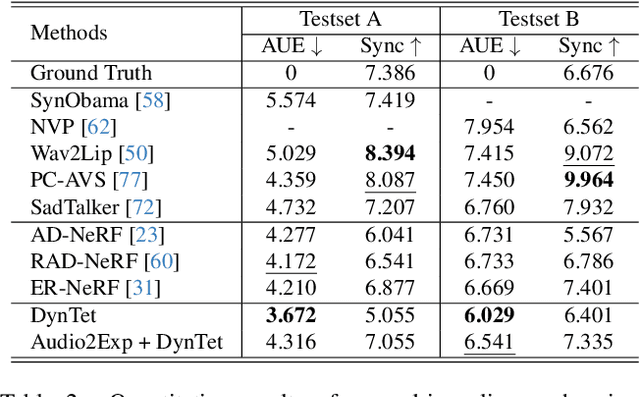
Abstract:Recent works in implicit representations, such as Neural Radiance Fields (NeRF), have advanced the generation of realistic and animatable head avatars from video sequences. These implicit methods are still confronted by visual artifacts and jitters, since the lack of explicit geometric constraints poses a fundamental challenge in accurately modeling complex facial deformations. In this paper, we introduce Dynamic Tetrahedra (DynTet), a novel hybrid representation that encodes explicit dynamic meshes by neural networks to ensure geometric consistency across various motions and viewpoints. DynTet is parameterized by the coordinate-based networks which learn signed distance, deformation, and material texture, anchoring the training data into a predefined tetrahedra grid. Leveraging Marching Tetrahedra, DynTet efficiently decodes textured meshes with a consistent topology, enabling fast rendering through a differentiable rasterizer and supervision via a pixel loss. To enhance training efficiency, we incorporate classical 3D Morphable Models to facilitate geometry learning and define a canonical space for simplifying texture learning. These advantages are readily achievable owing to the effective geometric representation employed in DynTet. Compared with prior works, DynTet demonstrates significant improvements in fidelity, lip synchronization, and real-time performance according to various metrics. Beyond producing stable and visually appealing synthesis videos, our method also outputs the dynamic meshes which is promising to enable many emerging applications.
Masked Reconstruction Contrastive Learning with Information Bottleneck Principle
Nov 15, 2022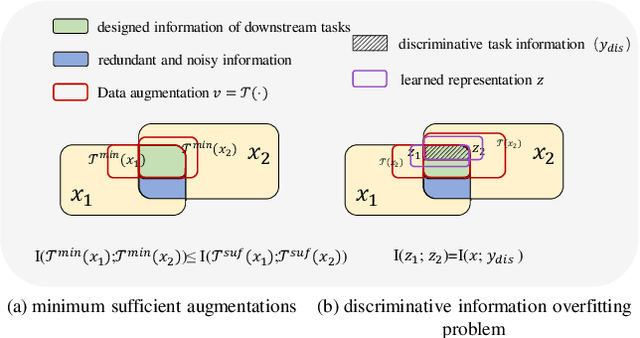
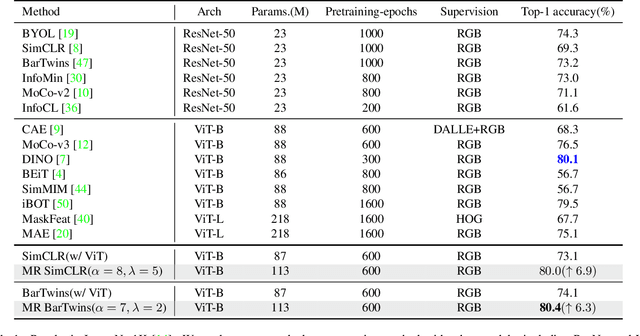
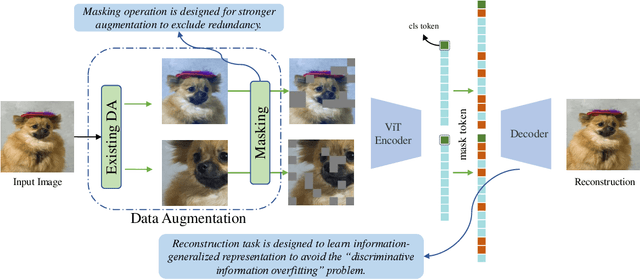
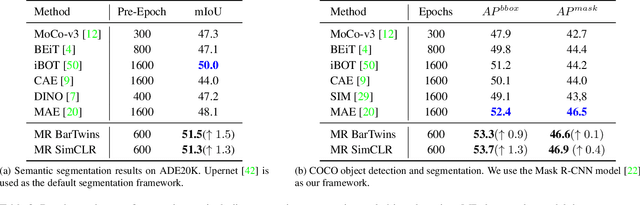
Abstract:Contrastive learning (CL) has shown great power in self-supervised learning due to its ability to capture insight correlations among large-scale data. Current CL models are biased to learn only the ability to discriminate positive and negative pairs due to the discriminative task setting. However, this bias would lead to ignoring its sufficiency for other downstream tasks, which we call the discriminative information overfitting problem. In this paper, we propose to tackle the above problems from the aspect of the Information Bottleneck (IB) principle, further pushing forward the frontier of CL. Specifically, we present a new perspective that CL is an instantiation of the IB principle, including information compression and expression. We theoretically analyze the optimal information situation and demonstrate that minimum sufficient augmentation and information-generalized representation are the optimal requirements for achieving maximum compression and generalizability to downstream tasks. Therefore, we propose the Masked Reconstruction Contrastive Learning~(MRCL) model to improve CL models. For implementation in practice, MRCL utilizes the masking operation for stronger augmentation, further eliminating redundant and noisy information. In order to alleviate the discriminative information overfitting problem effectively, we employ the reconstruction task to regularize the discriminative task. We conduct comprehensive experiments and show the superiority of the proposed model on multiple tasks, including image classification, semantic segmentation and objective detection.
BERT-Flow-VAE: A Weakly-supervised Model for Multi-Label Text Classification
Oct 27, 2022



Abstract:Multi-label Text Classification (MLTC) is the task of categorizing documents into one or more topics. Considering the large volumes of data and varying domains of such tasks, fully supervised learning requires manually fully annotated datasets which is costly and time-consuming. In this paper, we propose BERT-Flow-VAE (BFV), a Weakly-Supervised Multi-Label Text Classification (WSMLTC) model that reduces the need for full supervision. This new model (1) produces BERT sentence embeddings and calibrates them using a flow model, (2) generates an initial topic-document matrix by averaging results of a seeded sparse topic model and a textual entailment model which only require surface name of topics and 4-6 seed words per topic, and (3) adopts a VAE framework to reconstruct the embeddings under the guidance of the topic-document matrix. Finally, (4) it uses the means produced by the encoder model in the VAE architecture as predictions for MLTC. Experimental results on 6 multi-label datasets show that BFV can substantially outperform other baseline WSMLTC models in key metrics and achieve approximately 84% performance of a fully-supervised model.
* 8 pages, 4 figures
Graph Representation Learning by Ensemble Aggregating Subgraphs via Mutual Information Maximization
Mar 24, 2021



Abstract:Graph Neural Networks have shown tremendous potential on dealing with garph data and achieved outstanding results in recent years. In some research areas, labelling data are hard to obtain for technical reasons, which necessitates the study of unsupervised and semi-superivsed learning on graphs. Therefore, whether the learned representations can capture the intrinsic feature of the original graphs will be the issue in this area. In this paper, we introduce a self-supervised learning method to enhance the representations of graph-level learned by Graph Neural Networks. To fully capture the original attributes of the graph, we use three information aggregators: attribute-conv, layer-conv and subgraph-conv to gather information from different aspects. To get a comprehensive understanding of the graph structure, we propose an ensemble-learning like subgraph method. And to achieve efficient and effective contrasive learning, a Head-Tail contrastive samples construction method is proposed to provide more abundant negative samples. By virtue of all proposed components which can be generalized to any Graph Neural Networks, in unsupervised case, we achieve new state of the art results in several benchmarks. We also evaluate our model on semi-supervised learning tasks and make a fair comparison to state of the art semi-supervised methods.
Blocked and Hierarchical Disentangled Representation From Information Theory Perspective
Jan 21, 2021



Abstract:We propose a novel and theoretical model, blocked and hierarchical variational autoencoder (BHiVAE), to get better-disentangled representation. It is well known that information theory has an excellent explanatory meaning for the network, so we start to solve the disentanglement problem from the perspective of information theory. BHiVAE mainly comes from the information bottleneck theory and information maximization principle. Our main idea is that (1) Neurons block not only one neuron node is used to represent attribute, which can contain enough information; (2) Create a hierarchical structure with different attributes on different layers, so that we can segment the information within each layer to ensure that the final representation is disentangled. Furthermore, we present supervised and unsupervised BHiVAE, respectively, where the difference is mainly reflected in the separation of information between different blocks. In supervised BHiVAE, we utilize the label information as the standard to separate blocks. In unsupervised BHiVAE, without extra information, we use the Total Correlation (TC) measure to achieve independence, and we design a new prior distribution of the latent space to guide the representation learning. It also exhibits excellent disentanglement results in experiments and superior classification accuracy in representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge