Yanzhou Su
A Survey of Scientific Large Language Models: From Data Foundations to Agent Frontiers
Aug 28, 2025



Abstract:Scientific Large Language Models (Sci-LLMs) are transforming how knowledge is represented, integrated, and applied in scientific research, yet their progress is shaped by the complex nature of scientific data. This survey presents a comprehensive, data-centric synthesis that reframes the development of Sci-LLMs as a co-evolution between models and their underlying data substrate. We formulate a unified taxonomy of scientific data and a hierarchical model of scientific knowledge, emphasizing the multimodal, cross-scale, and domain-specific challenges that differentiate scientific corpora from general natural language processing datasets. We systematically review recent Sci-LLMs, from general-purpose foundations to specialized models across diverse scientific disciplines, alongside an extensive analysis of over 270 pre-/post-training datasets, showing why Sci-LLMs pose distinct demands -- heterogeneous, multi-scale, uncertainty-laden corpora that require representations preserving domain invariance and enabling cross-modal reasoning. On evaluation, we examine over 190 benchmark datasets and trace a shift from static exams toward process- and discovery-oriented assessments with advanced evaluation protocols. These data-centric analyses highlight persistent issues in scientific data development and discuss emerging solutions involving semi-automated annotation pipelines and expert validation. Finally, we outline a paradigm shift toward closed-loop systems where autonomous agents based on Sci-LLMs actively experiment, validate, and contribute to a living, evolving knowledge base. Collectively, this work provides a roadmap for building trustworthy, continually evolving artificial intelligence (AI) systems that function as a true partner in accelerating scientific discovery.
Enhancing LLMs' Reasoning-Intensive Multimedia Search Capabilities through Fine-Tuning and Reinforcement Learning
May 24, 2025Abstract:Existing large language models (LLMs) driven search agents typically rely on prompt engineering to decouple the user queries into search plans, limiting their effectiveness in complex scenarios requiring reasoning. Furthermore, they suffer from excessive token consumption due to Python-based search plan representations and inadequate integration of multimedia elements for both input processing and response generation. To address these challenges, we introduce SearchExpert, a training method for LLMs to improve their multimedia search capabilities in response to complex search queries. Firstly, we reformulate the search plan in an efficient natural language representation to reduce token consumption. Then, we propose the supervised fine-tuning for searching (SFTS) to fine-tune LLM to adapt to these representations, together with an automated dataset construction pipeline. Secondly, to improve reasoning-intensive search capabilities, we propose the reinforcement learning from search feedback (RLSF) that takes the search results planned by LLM as the reward signals. Thirdly, we propose a multimedia understanding and generation agent that enables the fine-tuned LLM to process visual input and produce visual output during inference. Finally, we establish an automated benchmark construction pipeline and a human evaluation framework. Our resulting benchmark, SearchExpertBench-25, comprises 200 multiple-choice questions spanning financial and international news scenarios that require reasoning in searching. Experiments demonstrate that SearchExpert outperforms the commercial LLM search method (Perplexity Pro) by 36.60% on the existing FinSearchBench-24 benchmark and 54.54% on our proposed SearchExpertBench-25. Human evaluations further confirm the superior readability.
RetinaLogos: Fine-Grained Synthesis of High-Resolution Retinal Images Through Captions
May 19, 2025Abstract:The scarcity of high-quality, labelled retinal imaging data, which presents a significant challenge in the development of machine learning models for ophthalmology, hinders progress in the field. To synthesise Colour Fundus Photographs (CFPs), existing methods primarily relying on predefined disease labels face significant limitations. However, current methods remain limited, thus failing to generate images for broader categories with diverse and fine-grained anatomical structures. To overcome these challenges, we first introduce an innovative pipeline that creates a large-scale, synthetic Caption-CFP dataset comprising 1.4 million entries, called RetinaLogos-1400k. Specifically, RetinaLogos-1400k uses large language models (LLMs) to describe retinal conditions and key structures, such as optic disc configuration, vascular distribution, nerve fibre layers, and pathological features. Furthermore, based on this dataset, we employ a novel three-step training framework, called RetinaLogos, which enables fine-grained semantic control over retinal images and accurately captures different stages of disease progression, subtle anatomical variations, and specific lesion types. Extensive experiments demonstrate state-of-the-art performance across multiple datasets, with 62.07% of text-driven synthetic images indistinguishable from real ones by ophthalmologists. Moreover, the synthetic data improves accuracy by 10%-25% in diabetic retinopathy grading and glaucoma detection, thereby providing a scalable solution to augment ophthalmic datasets.
GMAI-VL-R1: Harnessing Reinforcement Learning for Multimodal Medical Reasoning
Apr 02, 2025Abstract:Recent advances in general medical AI have made significant strides, but existing models often lack the reasoning capabilities needed for complex medical decision-making. This paper presents GMAI-VL-R1, a multimodal medical reasoning model enhanced by reinforcement learning (RL) to improve its reasoning abilities. Through iterative training, GMAI-VL-R1 optimizes decision-making, significantly boosting diagnostic accuracy and clinical support. We also develop a reasoning data synthesis method, generating step-by-step reasoning data via rejection sampling, which further enhances the model's generalization. Experimental results show that after RL training, GMAI-VL-R1 excels in tasks such as medical image diagnosis and visual question answering. While the model demonstrates basic memorization with supervised fine-tuning, RL is crucial for true generalization. Our work establishes new evaluation benchmarks and paves the way for future advancements in medical reasoning models. Code, data, and model will be released at \href{https://github.com/uni-medical/GMAI-VL-R1}{this link}.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025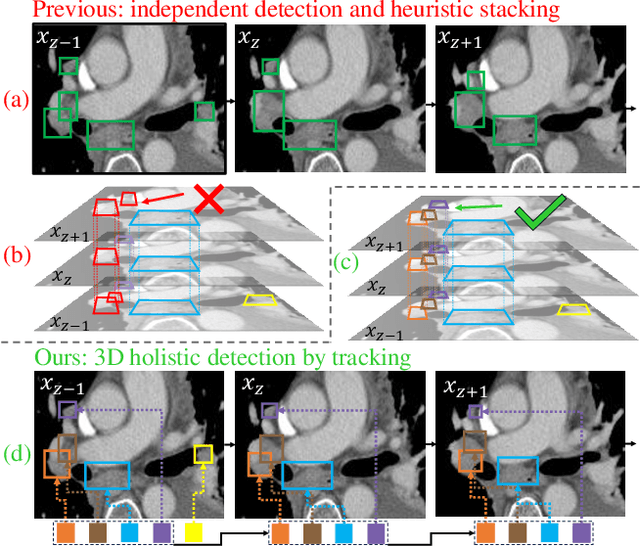

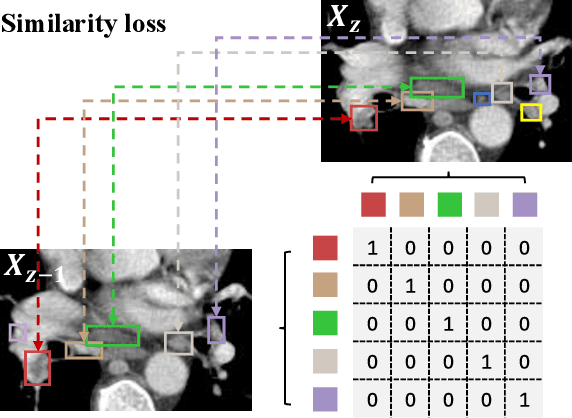
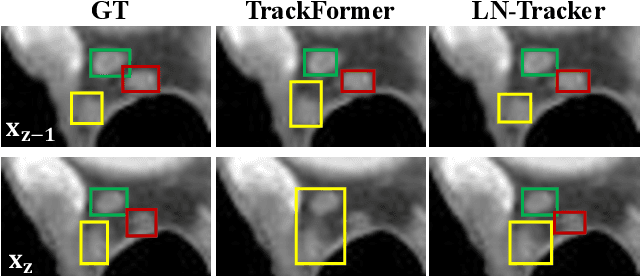
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
Towards Effective and Efficient Context-aware Nucleus Detection in Histopathology Whole Slide Images
Mar 04, 2025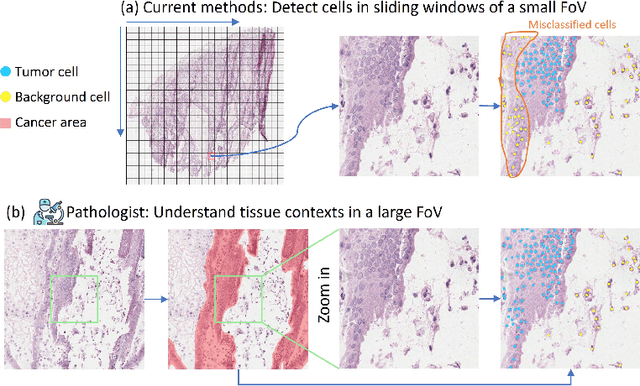
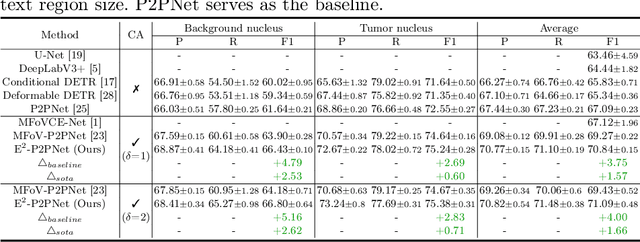
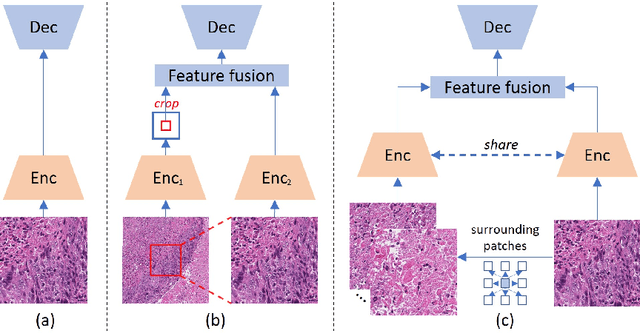
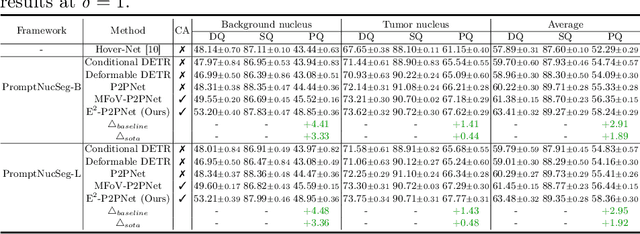
Abstract:Nucleus detection in histopathology whole slide images (WSIs) is crucial for a broad spectrum of clinical applications. Current approaches for nucleus detection in gigapixel WSIs utilize a sliding window methodology, which overlooks boarder contextual information (eg, tissue structure) and easily leads to inaccurate predictions. To address this problem, recent studies additionally crops a large Filed-of-View (FoV) region around each sliding window to extract contextual features. However, such methods substantially increases the inference latency. In this paper, we propose an effective and efficient context-aware nucleus detection algorithm. Specifically, instead of leveraging large FoV regions, we aggregate contextual clues from off-the-shelf features of historically visited sliding windows. This design greatly reduces computational overhead. Moreover, compared to large FoV regions at a low magnification, the sliding window patches have higher magnification and provide finer-grained tissue details, thereby enhancing the detection accuracy. To further improve the efficiency, we propose a grid pooling technique to compress dense feature maps of each patch into a few contextual tokens. Finally, we craft OCELOT-seg, the first benchmark dedicated to context-aware nucleus instance segmentation. Code, dataset, and model checkpoints will be available at https://github.com/windygoo/PathContext.
Leveraging Semantic Asymmetry for Precise Gross Tumor Volume Segmentation of Nasopharyngeal Carcinoma in Planning CT
Nov 27, 2024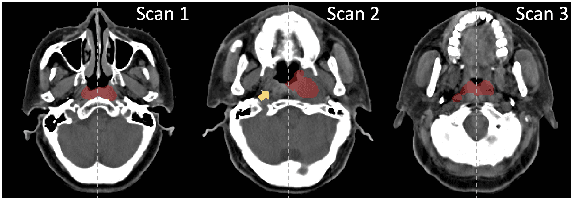
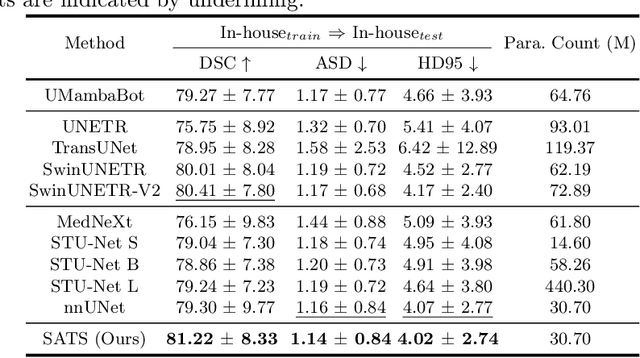
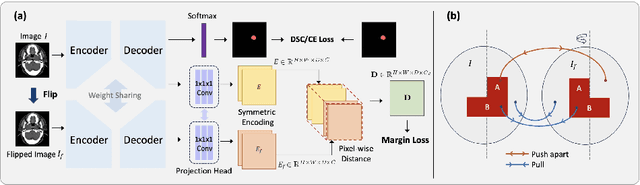
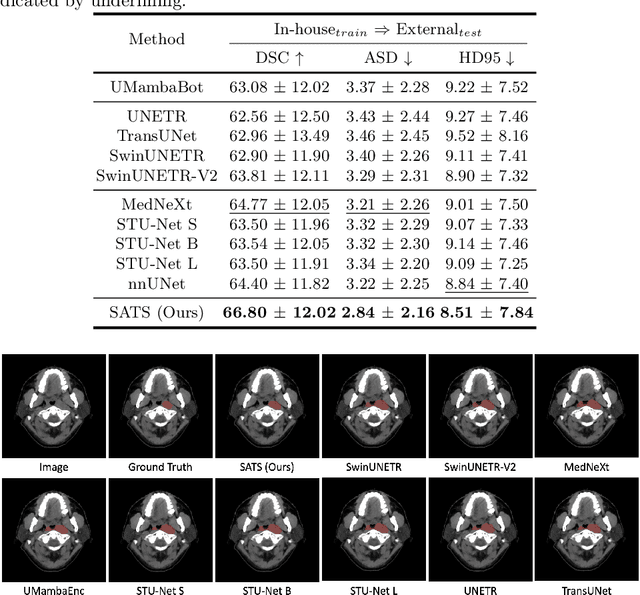
Abstract:In the radiation therapy of nasopharyngeal carcinoma (NPC), clinicians typically delineate the gross tumor volume (GTV) using non-contrast planning computed tomography to ensure accurate radiation dose delivery. However, the low contrast between tumors and adjacent normal tissues necessitates that radiation oncologists manually delineate the tumors, often relying on diagnostic MRI for guidance. % In this study, we propose a novel approach to directly segment NPC gross tumors on non-contrast planning CT images, circumventing potential registration errors when aligning MRI or MRI-derived tumor masks to planning CT. To address the low contrast issues between tumors and adjacent normal structures in planning CT, we introduce a 3D Semantic Asymmetry Tumor segmentation (SATs) method. Specifically, we posit that a healthy nasopharyngeal region is characteristically bilaterally symmetric, whereas the emergence of nasopharyngeal carcinoma disrupts this symmetry. Then, we propose a Siamese contrastive learning segmentation framework that minimizes the voxel-wise distance between original and flipped areas without tumor and encourages a larger distance between original and flipped areas with tumor. Thus, our approach enhances the sensitivity of features to semantic asymmetries. % Extensive experiments demonstrate that the proposed SATs achieves the leading NPC GTV segmentation performance in both internal and external testing, \emph{e.g.}, with at least 2\% absolute Dice score improvement and 12\% average distance error reduction when compared to other state-of-the-art methods in the external testing.
GMAI-VL & GMAI-VL-5.5M: A Large Vision-Language Model and A Comprehensive Multimodal Dataset Towards General Medical AI
Nov 21, 2024Abstract:Despite significant advancements in general artificial intelligence, such as GPT-4, their effectiveness in the medical domain (general medical AI, GMAI) remains constrained due to the absence of specialized medical knowledge. To address this challenge, we present GMAI-VL-5.5M, a comprehensive multimodal medical dataset created by converting hundreds of specialized medical datasets into meticulously constructed image-text pairs. This dataset features comprehensive task coverage, diverse modalities, and high-quality image-text data. Building upon this multimodal dataset, we propose GMAI-VL, a general medical vision-language model with a progressively three-stage training strategy. This approach significantly enhances the model's ability by integrating visual and textual information, thereby improving its ability to process multimodal data and support accurate diagnosis and clinical decision-making. Experimental evaluations demonstrate that GMAI-VL achieves state-of-the-art results across a wide range of multimodal medical tasks, such as visual question answering and medical image diagnosis. Our contributions include the development of the GMAI-VL-5.5M dataset, the introduction of the GMAI-VL model, and the establishment of new benchmarks in multiple medical domains. Code and dataset will be released at https://github.com/uni-medical/GMAI-VL.
SegBook: A Simple Baseline and Cookbook for Volumetric Medical Image Segmentation
Nov 21, 2024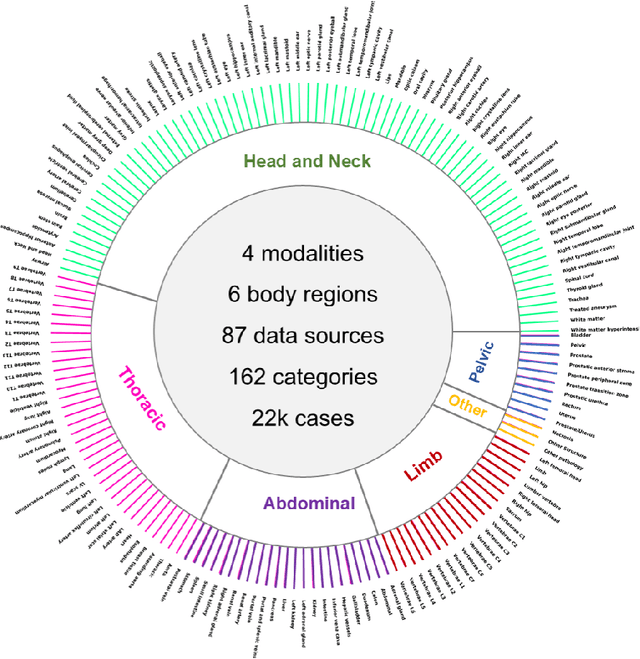
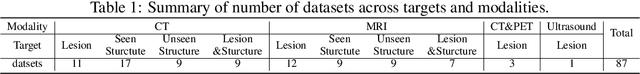
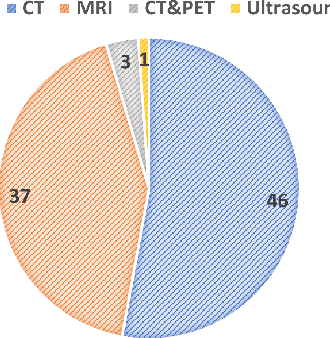
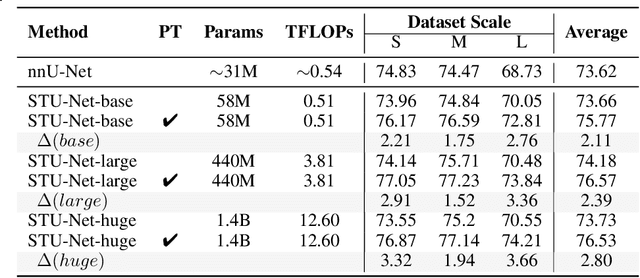
Abstract:Computed Tomography (CT) is one of the most popular modalities for medical imaging. By far, CT images have contributed to the largest publicly available datasets for volumetric medical segmentation tasks, covering full-body anatomical structures. Large amounts of full-body CT images provide the opportunity to pre-train powerful models, e.g., STU-Net pre-trained in a supervised fashion, to segment numerous anatomical structures. However, it remains unclear in which conditions these pre-trained models can be transferred to various downstream medical segmentation tasks, particularly segmenting the other modalities and diverse targets. To address this problem, a large-scale benchmark for comprehensive evaluation is crucial for finding these conditions. Thus, we collected 87 public datasets varying in modality, target, and sample size to evaluate the transfer ability of full-body CT pre-trained models. We then employed a representative model, STU-Net with multiple model scales, to conduct transfer learning across modalities and targets. Our experimental results show that (1) there may be a bottleneck effect concerning the dataset size in fine-tuning, with more improvement on both small- and large-scale datasets than medium-size ones. (2) Models pre-trained on full-body CT demonstrate effective modality transfer, adapting well to other modalities such as MRI. (3) Pre-training on the full-body CT not only supports strong performance in structure detection but also shows efficacy in lesion detection, showcasing adaptability across target tasks. We hope that this large-scale open evaluation of transfer learning can direct future research in volumetric medical image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge