Jiatong Cai
Towards Effective and Efficient Context-aware Nucleus Detection in Histopathology Whole Slide Images
Mar 04, 2025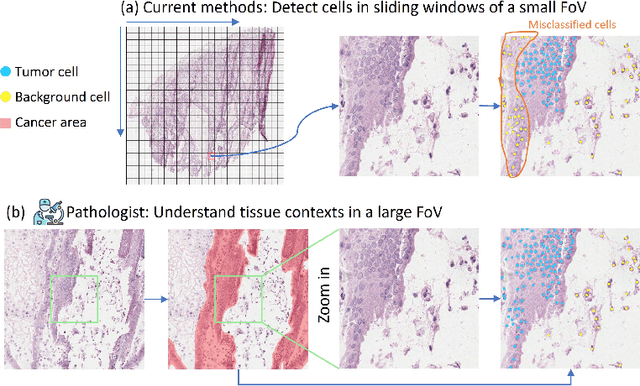
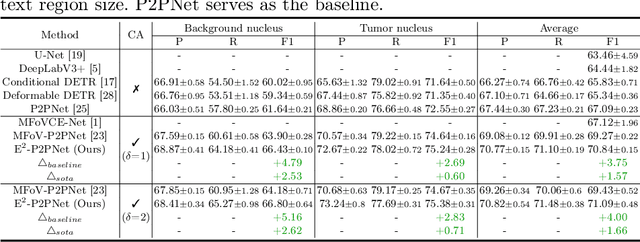
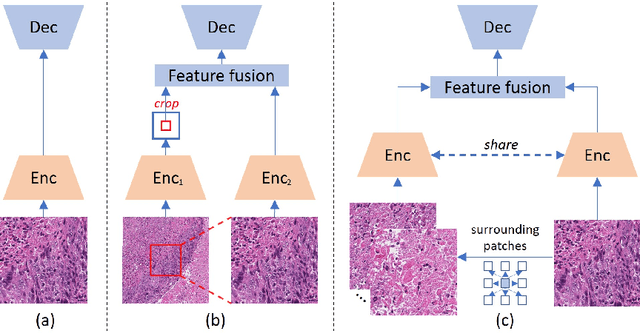
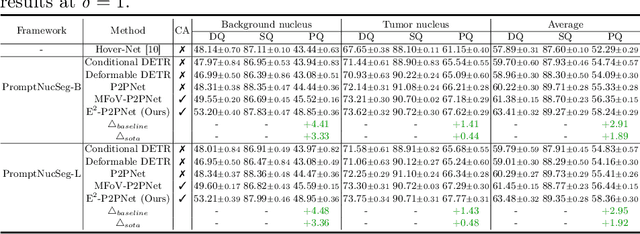
Abstract:Nucleus detection in histopathology whole slide images (WSIs) is crucial for a broad spectrum of clinical applications. Current approaches for nucleus detection in gigapixel WSIs utilize a sliding window methodology, which overlooks boarder contextual information (eg, tissue structure) and easily leads to inaccurate predictions. To address this problem, recent studies additionally crops a large Filed-of-View (FoV) region around each sliding window to extract contextual features. However, such methods substantially increases the inference latency. In this paper, we propose an effective and efficient context-aware nucleus detection algorithm. Specifically, instead of leveraging large FoV regions, we aggregate contextual clues from off-the-shelf features of historically visited sliding windows. This design greatly reduces computational overhead. Moreover, compared to large FoV regions at a low magnification, the sliding window patches have higher magnification and provide finer-grained tissue details, thereby enhancing the detection accuracy. To further improve the efficiency, we propose a grid pooling technique to compress dense feature maps of each patch into a few contextual tokens. Finally, we craft OCELOT-seg, the first benchmark dedicated to context-aware nucleus instance segmentation. Code, dataset, and model checkpoints will be available at https://github.com/windygoo/PathContext.
Long-MIL: Scaling Long Contextual Multiple Instance Learning for Histopathology Whole Slide Image Analysis
Nov 21, 2023Abstract:Histopathology image analysis is the golden standard of clinical diagnosis for Cancers. In doctors daily routine and computer-aided diagnosis, the Whole Slide Image (WSI) of histopathology tissue is used for analysis. Because of the extremely large scale of resolution, previous methods generally divide the WSI into a large number of patches, then aggregate all patches within a WSI by Multi-Instance Learning (MIL) to make the slide-level prediction when developing computer-aided diagnosis tools. However, most previous WSI-MIL models using global-attention without pairwise interaction and any positional information, or self-attention with absolute position embedding can not well handle shape varying large WSIs, e.g. testing WSIs after model deployment may be larger than training WSIs, since the model development set is always limited due to the difficulty of histopathology WSIs collection. To deal with the problem, in this paper, we propose to amend position embedding for shape varying long-contextual WSI by introducing Linear Bias into Attention, and adapt it from 1-d long sequence into 2-d long-contextual WSI which helps model extrapolate position embedding to unseen or under-fitted positions. We further utilize Flash-Attention module to tackle the computational complexity of Transformer, which also keep full self-attention performance compared to previous attention approximation work. Our method, Long-contextual MIL (Long-MIL) are evaluated on extensive experiments including 4 dataset including WSI classification and survival prediction tasks to validate the superiority on shape varying WSIs. The source code will be open-accessed soon.
Exploring Unsupervised Cell Recognition with Prior Self-activation Maps
Aug 22, 2023



Abstract:The success of supervised deep learning models on cell recognition tasks relies on detailed annotations. Many previous works have managed to reduce the dependency on labels. However, considering the large number of cells contained in a patch, costly and inefficient labeling is still inevitable. To this end, we explored label-free methods for cell recognition. Prior self-activation maps (PSM) are proposed to generate pseudo masks as training targets. To be specific, an activation network is trained with self-supervised learning. The gradient information in the shallow layers of the network is aggregated to generate prior self-activation maps. Afterward, a semantic clustering module is then introduced as a pipeline to transform PSMs to pixel-level semantic pseudo masks for downstream tasks. We evaluated our method on two histological datasets: MoNuSeg (cell segmentation) and BCData (multi-class cell detection). Compared with other fully-supervised and weakly-supervised methods, our method can achieve competitive performance without any manual annotations. Our simple but effective framework can also achieve multi-class cell detection which can not be done by existing unsupervised methods. The results show the potential of PSMs that might inspire other research to deal with the hunger for labels in medical area.
Unsupervised Dense Nuclei Detection and Segmentation with Prior Self-activation Map For Histology Images
Oct 14, 2022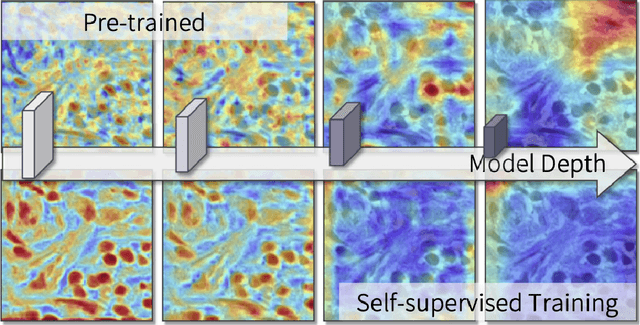
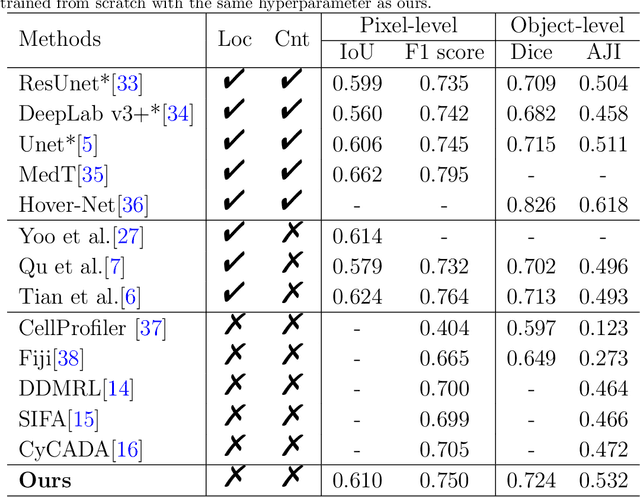
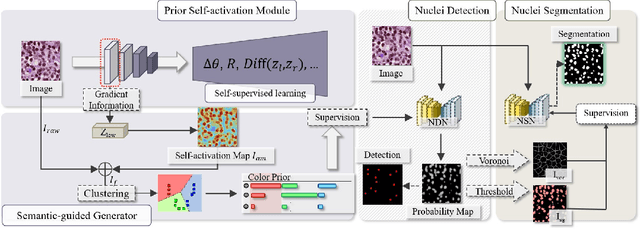
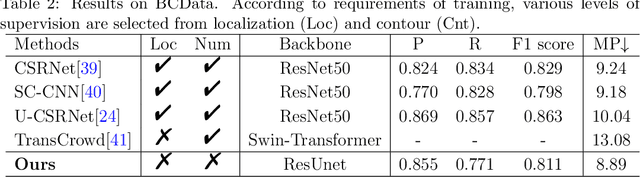
Abstract:The success of supervised deep learning models in medical image segmentation relies on detailed annotations. However, labor-intensive manual labeling is costly and inefficient, especially in dense object segmentation. To this end, we propose a self-supervised learning based approach with a Prior Self-activation Module (PSM) that generates self-activation maps from the input images to avoid labeling costs and further produce pseudo masks for the downstream task. To be specific, we firstly train a neural network using self-supervised learning and utilize the gradient information in the shallow layers of the network to generate self-activation maps. Afterwards, a semantic-guided generator is then introduced as a pipeline to transform visual representations from PSM to pixel-level semantic pseudo masks for downstream tasks. Furthermore, a two-stage training module, consisting of a nuclei detection network and a nuclei segmentation network, is adopted to achieve the final segmentation. Experimental results show the effectiveness on two public pathological datasets. Compared with other fully-supervised and weakly-supervised methods, our method can achieve competitive performance without any manual annotations.
Weakly Supervised Learning for cell recognition in immunohistochemical cytoplasm staining images
Feb 27, 2022



Abstract:Cell classification and counting in immunohistochemical cytoplasm staining images play a pivotal role in cancer diagnosis. Weakly supervised learning is a potential method to deal with labor-intensive labeling. However, the inconstant cell morphology and subtle differences between classes also bring challenges. To this end, we present a novel cell recognition framework based on multi-task learning, which utilizes two additional auxiliary tasks to guide robust representation learning of the main task. To deal with misclassification, the tissue prior learning branch is introduced to capture the spatial representation of tumor cells without additional tissue annotation. Moreover, dynamic masks and consistency learning are adopted to learn the invariance of cell scale and shape. We have evaluated our framework on immunohistochemical cytoplasm staining images, and the results demonstrate that our method outperforms recent cell recognition approaches. Besides, we have also done some ablation studies to show significant improvements after adding the auxiliary branches.
Generalizing Nucleus Recognition Model in Multi-source Images via Pruning
Jul 06, 2021



Abstract:Ki67 is a significant biomarker in the diagnosis and prognosis of cancer, whose index can be evaluated by quantifying its expression in Ki67 immunohistochemistry (IHC) stained images. However, quantitative analysis on multi-source Ki67 images is yet a challenging task in practice due to cross-domain distribution differences, which result from imaging variation, staining styles, and lesion types. Many recent studies have made some efforts on domain generalization (DG), whereas there are still some noteworthy limitations. Specifically in the case of Ki67 images, learning invariant representation is at the mercy of the insufficient number of domains and the cell categories mismatching in different domains. In this paper, we propose a novel method to improve DG by searching the domain-agnostic subnetwork in a domain merging scenario. Partial model parameters are iteratively pruned according to the domain gap, which is caused by the data converting from a single domain into merged domains during training. In addition, the model is optimized by fine-tuning on merged domains to eliminate the interference of class mismatching among various domains. Furthermore, an appropriate implementation is attained by applying the pruning method to different parts of the framework. Compared with known DG methods, our method yields excellent performance in multiclass nucleus recognition of Ki67 IHC images, especially in the lost category cases. Moreover, our competitive results are also evaluated on the public dataset over the state-of-the-art DG methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge