Can Cui
MULTISPEECH
FSDAM: Few-Shot Driving Attention Modeling via Vision-Language Coupling
Nov 16, 2025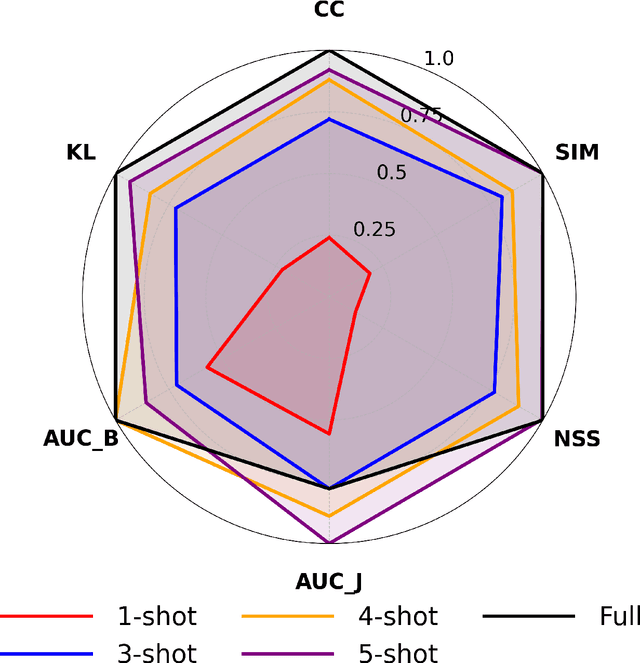
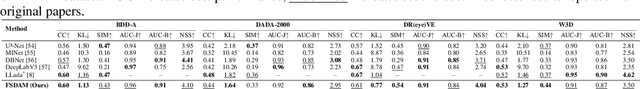
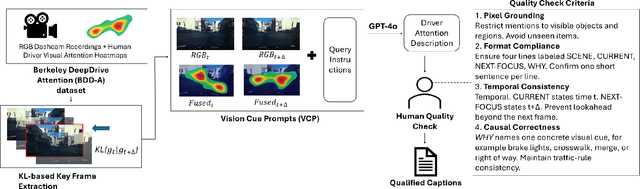
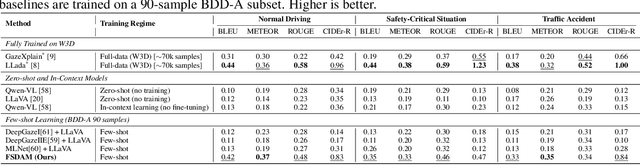
Abstract:Understanding where drivers look and why they shift their attention is essential for autonomous systems that read human intent and justify their actions. Most existing models rely on large-scale gaze datasets to learn these patterns; however, such datasets are labor-intensive to collect and time-consuming to curate. We present FSDAM (Few-Shot Driver Attention Modeling), a framework that achieves joint attention prediction and caption generation with approximately 100 annotated examples, two orders of magnitude fewer than existing approaches. Our approach introduces a dual-pathway architecture where separate modules handle spatial prediction and caption generation while maintaining semantic consistency through cross-modal alignment. Despite minimal supervision, FSDAM achieves competitive performance on attention prediction, generates coherent, and context-aware explanations. The model demonstrates robust zero-shot generalization across multiple driving benchmarks. This work shows that effective attention-conditioned generation is achievable with limited supervision, opening new possibilities for practical deployment of explainable driver attention systems in data-constrained scenarios.
VLA-Adapter: An Effective Paradigm for Tiny-Scale Vision-Language-Action Model
Sep 11, 2025Abstract:Vision-Language-Action (VLA) models typically bridge the gap between perceptual and action spaces by pre-training a large-scale Vision-Language Model (VLM) on robotic data. While this approach greatly enhances performance, it also incurs significant training costs. In this paper, we investigate how to effectively bridge vision-language (VL) representations to action (A). We introduce VLA-Adapter, a novel paradigm designed to reduce the reliance of VLA models on large-scale VLMs and extensive pre-training. To this end, we first systematically analyze the effectiveness of various VL conditions and present key findings on which conditions are essential for bridging perception and action spaces. Based on these insights, we propose a lightweight Policy module with Bridge Attention, which autonomously injects the optimal condition into the action space. In this way, our method achieves high performance using only a 0.5B-parameter backbone, without any robotic data pre-training. Extensive experiments on both simulated and real-world robotic benchmarks demonstrate that VLA-Adapter not only achieves state-of-the-art level performance, but also offers the fast inference speed reported to date. Furthermore, thanks to the proposed advanced bridging paradigm, VLA-Adapter enables the training of a powerful VLA model in just 8 hours on a single consumer-grade GPU, greatly lowering the barrier to deploying the VLA model. Project page: https://vla-adapter.github.io/.
ViLaD: A Large Vision Language Diffusion Framework for End-to-End Autonomous Driving
Aug 18, 2025Abstract:End-to-end autonomous driving systems built on Vision Language Models (VLMs) have shown significant promise, yet their reliance on autoregressive architectures introduces some limitations for real-world applications. The sequential, token-by-token generation process of these models results in high inference latency and cannot perform bidirectional reasoning, making them unsuitable for dynamic, safety-critical environments. To overcome these challenges, we introduce ViLaD, a novel Large Vision Language Diffusion (LVLD) framework for end-to-end autonomous driving that represents a paradigm shift. ViLaD leverages a masked diffusion model that enables parallel generation of entire driving decision sequences, significantly reducing computational latency. Moreover, its architecture supports bidirectional reasoning, allowing the model to consider both past and future simultaneously, and supports progressive easy-first generation to iteratively improve decision quality. We conduct comprehensive experiments on the nuScenes dataset, where ViLaD outperforms state-of-the-art autoregressive VLM baselines in both planning accuracy and inference speed, while achieving a near-zero failure rate. Furthermore, we demonstrate the framework's practical viability through a real-world deployment on an autonomous vehicle for an interactive parking task, confirming its effectiveness and soundness for practical applications.
Quantitative Benchmarking of Anomaly Detection Methods in Digital Pathology
Jun 24, 2025Abstract:Anomaly detection has been widely studied in the context of industrial defect inspection, with numerous methods developed to tackle a range of challenges. In digital pathology, anomaly detection holds significant potential for applications such as rare disease identification, artifact detection, and biomarker discovery. However, the unique characteristics of pathology images, such as their large size, multi-scale structures, stain variability, and repetitive patterns, introduce new challenges that current anomaly detection algorithms struggle to address. In this quantitative study, we benchmark over 20 classical and prevalent anomaly detection methods through extensive experiments. We curated five digital pathology datasets, both real and synthetic, to systematically evaluate these approaches. Our experiments investigate the influence of image scale, anomaly pattern types, and training epoch selection strategies on detection performance. The results provide a detailed comparison of each method's strengths and limitations, establishing a comprehensive benchmark to guide future research in anomaly detection for digital pathology images.
A Hierarchical Test Platform for Vision Language Model (VLM)-Integrated Real-World Autonomous Driving
Jun 17, 2025Abstract:Vision-Language Models (VLMs) have demonstrated notable promise in autonomous driving by offering the potential for multimodal reasoning through pretraining on extensive image-text pairs. However, adapting these models from broad web-scale data to the safety-critical context of driving presents a significant challenge, commonly referred to as domain shift. Existing simulation-based and dataset-driven evaluation methods, although valuable, often fail to capture the full complexity of real-world scenarios and cannot easily accommodate repeatable closed-loop testing with flexible scenario manipulation. In this paper, we introduce a hierarchical real-world test platform specifically designed to evaluate VLM-integrated autonomous driving systems. Our approach includes a modular, low-latency on-vehicle middleware that allows seamless incorporation of various VLMs, a clearly separated perception-planning-control architecture that can accommodate both VLM-based and conventional modules, and a configurable suite of real-world testing scenarios on a closed track that facilitates controlled yet authentic evaluations. We demonstrate the effectiveness of the proposed platform`s testing and evaluation ability with a case study involving a VLM-enabled autonomous vehicle, highlighting how our test framework supports robust experimentation under diverse conditions.
IRS: Incremental Relationship-guided Segmentation for Digital Pathology
May 28, 2025Abstract:Continual learning is rapidly emerging as a key focus in computer vision, aiming to develop AI systems capable of continuous improvement, thereby enhancing their value and practicality in diverse real-world applications. In healthcare, continual learning holds great promise for continuously acquired digital pathology data, which is collected in hospitals on a daily basis. However, panoramic segmentation on digital whole slide images (WSIs) presents significant challenges, as it is often infeasible to obtain comprehensive annotations for all potential objects, spanning from coarse structures (e.g., regions and unit objects) to fine structures (e.g., cells). This results in temporally and partially annotated data, posing a major challenge in developing a holistic segmentation framework. Moreover, an ideal segmentation model should incorporate new phenotypes, unseen diseases, and diverse populations, making this task even more complex. In this paper, we introduce a novel and unified Incremental Relationship-guided Segmentation (IRS) learning scheme to address temporally acquired, partially annotated data while maintaining out-of-distribution (OOD) continual learning capacity in digital pathology. The key innovation of IRS lies in its ability to realize a new spatial-temporal OOD continual learning paradigm by mathematically modeling anatomical relationships between existing and newly introduced classes through a simple incremental universal proposition matrix. Experimental results demonstrate that the IRS method effectively handles the multi-scale nature of pathological segmentation, enabling precise kidney segmentation across various structures (regions, units, and cells) as well as OOD disease lesions at multiple magnifications. This capability significantly enhances domain generalization, making IRS a robust approach for real-world digital pathology applications.
OpenHelix: A Short Survey, Empirical Analysis, and Open-Source Dual-System VLA Model for Robotic Manipulation
May 06, 2025



Abstract:Dual-system VLA (Vision-Language-Action) architectures have become a hot topic in embodied intelligence research, but there is a lack of sufficient open-source work for further performance analysis and optimization. To address this problem, this paper will summarize and compare the structural designs of existing dual-system architectures, and conduct systematic empirical evaluations on the core design elements of existing dual-system architectures. Ultimately, it will provide a low-cost open-source model for further exploration. Of course, this project will continue to update with more experimental conclusions and open-source models with improved performance for everyone to choose from. Project page: https://openhelix-robot.github.io/.
DeepAndes: A Self-Supervised Vision Foundation Model for Multi-Spectral Remote Sensing Imagery of the Andes
Apr 28, 2025Abstract:By mapping sites at large scales using remotely sensed data, archaeologists can generate unique insights into long-term demographic trends, inter-regional social networks, and past adaptations to climate change. Remote sensing surveys complement field-based approaches, and their reach can be especially great when combined with deep learning and computer vision techniques. However, conventional supervised deep learning methods face challenges in annotating fine-grained archaeological features at scale. While recent vision foundation models have shown remarkable success in learning large-scale remote sensing data with minimal annotations, most off-the-shelf solutions are designed for RGB images rather than multi-spectral satellite imagery, such as the 8-band data used in our study. In this paper, we introduce DeepAndes, a transformer-based vision foundation model trained on three million multi-spectral satellite images, specifically tailored for Andean archaeology. DeepAndes incorporates a customized DINOv2 self-supervised learning algorithm optimized for 8-band multi-spectral imagery, marking the first foundation model designed explicitly for the Andes region. We evaluate its image understanding performance through imbalanced image classification, image instance retrieval, and pixel-level semantic segmentation tasks. Our experiments show that DeepAndes achieves superior F1 scores, mean average precision, and Dice scores in few-shot learning scenarios, significantly outperforming models trained from scratch or pre-trained on smaller datasets. This underscores the effectiveness of large-scale self-supervised pre-training in archaeological remote sensing. Codes will be available on https://github.com/geopacha/DeepAndes.
NuPlanQA: A Large-Scale Dataset and Benchmark for Multi-View Driving Scene Understanding in Multi-Modal Large Language Models
Mar 17, 2025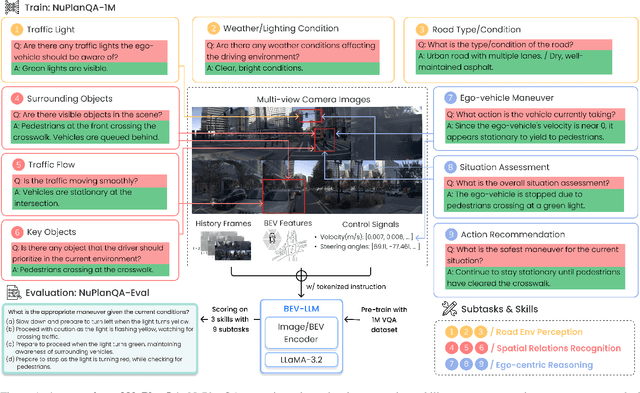
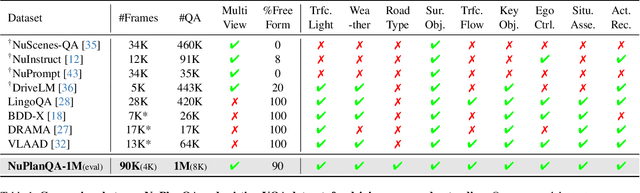
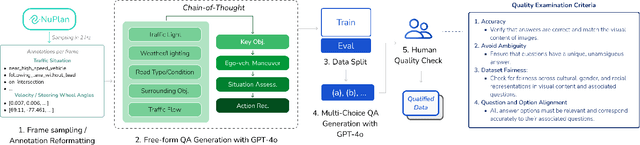
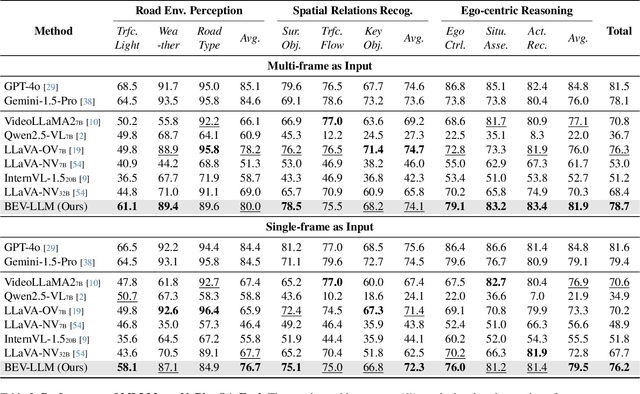
Abstract:Recent advances in multi-modal large language models (MLLMs) have demonstrated strong performance across various domains; however, their ability to comprehend driving scenes remains less proven. The complexity of driving scenarios, which includes multi-view information, poses significant challenges for existing MLLMs. In this paper, we introduce NuPlanQA-Eval, a multi-view, multi-modal evaluation benchmark for driving scene understanding. To further support generalization to multi-view driving scenarios, we also propose NuPlanQA-1M, a large-scale dataset comprising 1M real-world visual question-answering (VQA) pairs. For context-aware analysis of traffic scenes, we categorize our dataset into nine subtasks across three core skills: Road Environment Perception, Spatial Relations Recognition, and Ego-Centric Reasoning. Furthermore, we present BEV-LLM, integrating Bird's-Eye-View (BEV) features from multi-view images into MLLMs. Our evaluation results reveal key challenges that existing MLLMs face in driving scene-specific perception and spatial reasoning from ego-centric perspectives. In contrast, BEV-LLM demonstrates remarkable adaptability to this domain, outperforming other models in six of the nine subtasks. These findings highlight how BEV integration enhances multi-view MLLMs while also identifying key areas that require further refinement for effective adaptation to driving scenes. To facilitate further research, we publicly release NuPlanQA at https://github.com/sungyeonparkk/NuPlanQA.
QUART-Online: Latency-Free Large Multimodal Language Model for Quadruped Robot Learning
Dec 23, 2024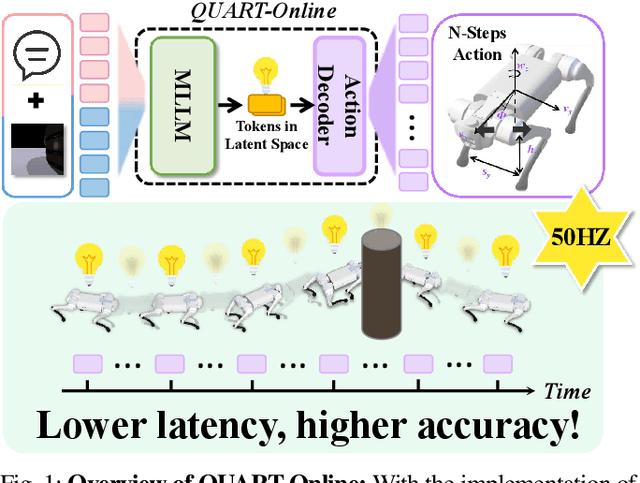
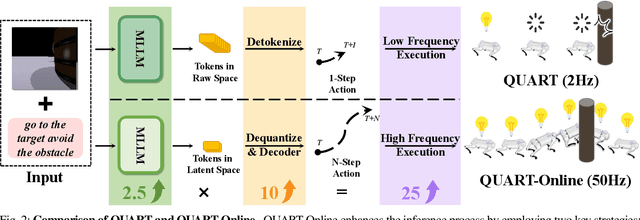
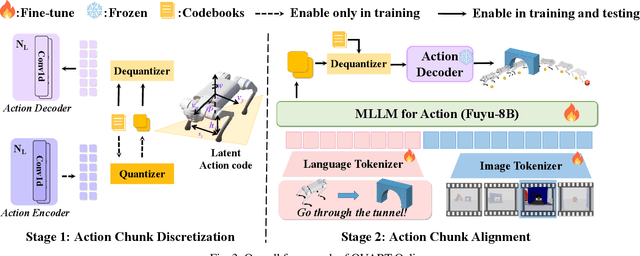
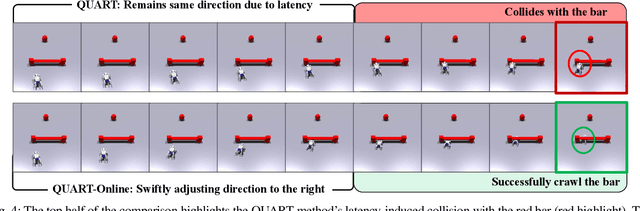
Abstract:This paper addresses the inherent inference latency challenges associated with deploying multimodal large language models (MLLM) in quadruped vision-language-action (QUAR-VLA) tasks. Our investigation reveals that conventional parameter reduction techniques ultimately impair the performance of the language foundation model during the action instruction tuning phase, making them unsuitable for this purpose. We introduce a novel latency-free quadruped MLLM model, dubbed QUART-Online, designed to enhance inference efficiency without degrading the performance of the language foundation model. By incorporating Action Chunk Discretization (ACD), we compress the original action representation space, mapping continuous action values onto a smaller set of discrete representative vectors while preserving critical information. Subsequently, we fine-tune the MLLM to integrate vision, language, and compressed actions into a unified semantic space. Experimental results demonstrate that QUART-Online operates in tandem with the existing MLLM system, achieving real-time inference in sync with the underlying controller frequency, significantly boosting the success rate across various tasks by 65%. Our project page is https://quart-online.github.io.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge