Jialin Yue
HistoWAS: A Pathomics Framework for Large-Scale Feature-Wide Association Studies of Tissue Topology and Patient Outcomes
Dec 23, 2025Abstract:High-throughput "pathomic" analysis of Whole Slide Images (WSIs) offers new opportunities to study tissue characteristics and for biomarker discovery. However, the clinical relevance of the tissue characteristics at the micro- and macro-environment level is limited by the lack of tools that facilitate the measurement of the spatial interaction of individual structure characteristics and their association with clinical parameters. To address these challenges, we introduce HistoWAS (Histology-Wide Association Study), a computational framework designed to link tissue spatial organization to clinical outcomes. Specifically, HistoWAS implements (1) a feature space that augments conventional metrics with 30 topological and spatial features, adapted from Geographic Information Systems (GIS) point pattern analysis, to quantify tissue micro-architecture; and (2) an association study engine, inspired by Phenome-Wide Association Studies (PheWAS), that performs mass univariate regression for each feature with statistical correction. As a proof of concept, we applied HistoWAS to analyze a total of 102 features (72 conventional object-level features and our 30 spatial features) using 385 PAS-stained WSIs from 206 participants in the Kidney Precision Medicine Project (KPMP). The code and data have been released to https://github.com/hrlblab/histoWAS.
DeepAndes: A Self-Supervised Vision Foundation Model for Multi-Spectral Remote Sensing Imagery of the Andes
Apr 28, 2025Abstract:By mapping sites at large scales using remotely sensed data, archaeologists can generate unique insights into long-term demographic trends, inter-regional social networks, and past adaptations to climate change. Remote sensing surveys complement field-based approaches, and their reach can be especially great when combined with deep learning and computer vision techniques. However, conventional supervised deep learning methods face challenges in annotating fine-grained archaeological features at scale. While recent vision foundation models have shown remarkable success in learning large-scale remote sensing data with minimal annotations, most off-the-shelf solutions are designed for RGB images rather than multi-spectral satellite imagery, such as the 8-band data used in our study. In this paper, we introduce DeepAndes, a transformer-based vision foundation model trained on three million multi-spectral satellite images, specifically tailored for Andean archaeology. DeepAndes incorporates a customized DINOv2 self-supervised learning algorithm optimized for 8-band multi-spectral imagery, marking the first foundation model designed explicitly for the Andes region. We evaluate its image understanding performance through imbalanced image classification, image instance retrieval, and pixel-level semantic segmentation tasks. Our experiments show that DeepAndes achieves superior F1 scores, mean average precision, and Dice scores in few-shot learning scenarios, significantly outperforming models trained from scratch or pre-trained on smaller datasets. This underscores the effectiveness of large-scale self-supervised pre-training in archaeological remote sensing. Codes will be available on https://github.com/geopacha/DeepAndes.
GloFinder: AI-empowered QuPath Plugin for WSI-level Glomerular Detection, Visualization, and Curation
Nov 27, 2024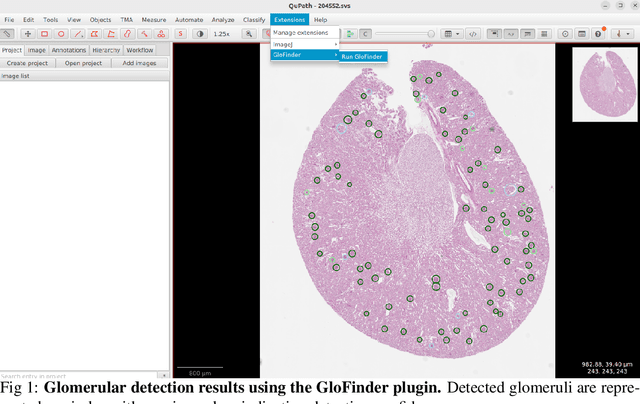
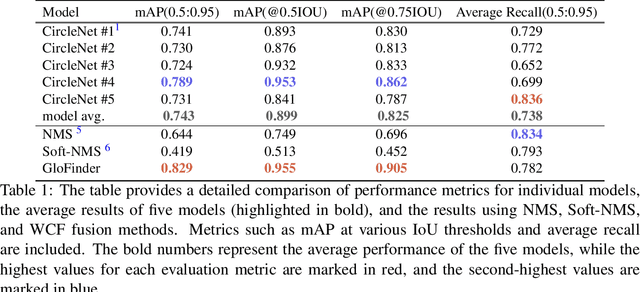
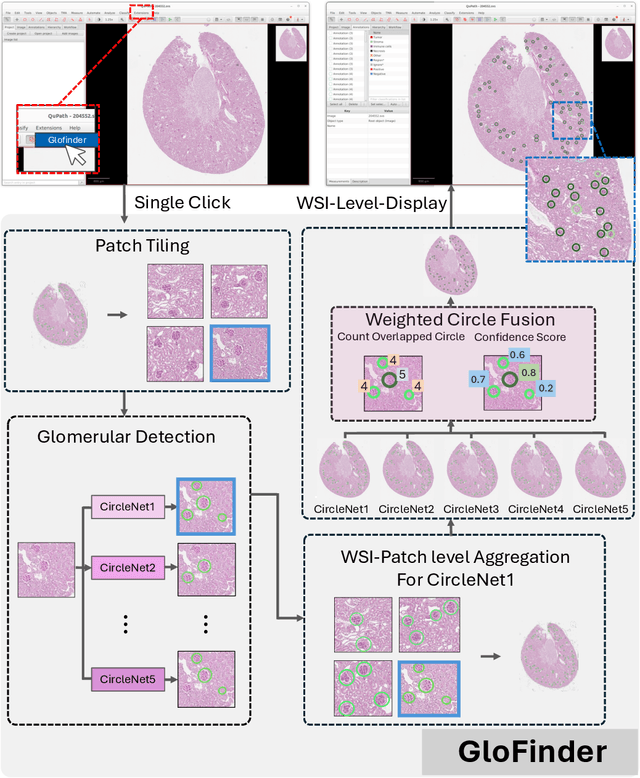
Abstract:Artificial intelligence (AI) has demonstrated significant success in automating the detection of glomeruli, the key functional units of the kidney, from whole slide images (WSIs) in kidney pathology. However, existing open-source tools are often distributed as source code or Docker containers, requiring advanced programming skills that hinder accessibility for non-programmers, such as clinicians. Additionally, current models are typically trained on a single dataset and lack flexibility in adjusting confidence levels for predictions. To overcome these challenges, we introduce GloFinder, a QuPath plugin designed for single-click automated glomeruli detection across entire WSIs with online editing through the graphical user interface (GUI). GloFinder employs CircleNet, an anchor-free detection framework utilizing circle representations for precise object localization, with models trained on approximately 160,000 manually annotated glomeruli. To further enhance accuracy, the plugin incorporates Weighted Circle Fusion (WCF), an ensemble method that combines confidence scores from multiple CircleNet models to produce refined predictions, achieving superior performance in glomerular detection. GloFinder enables direct visualization and editing of results in QuPath, facilitating seamless interaction for clinicians and providing a powerful tool for nephropathology research and clinical practice.
Cross-organ Deployment of EOS Detection AI without Retraining: Feasibility and Limitation
Nov 24, 2024



Abstract:Chronic rhinosinusitis (CRS) is characterized by persistent inflammation in the paranasal sinuses, leading to typical symptoms of nasal congestion, facial pressure, olfactory dysfunction, and discolored nasal drainage, which can significantly impact quality-of-life. Eosinophils (Eos), a crucial component in the mucosal immune response, have been linked to disease severity in CRS. The diagnosis of eosinophilic CRS typically uses a threshold of 10-20 eos per high-power field (HPF). However, manually counting Eos in histological samples is laborious and time-intensive, making the use of AI-driven methods for automated evaluations highly desirable. Interestingly, eosinophils are predominantly located in the gastrointestinal (GI) tract, which has prompted the release of numerous deep learning models trained on GI data. This study leverages a CircleSnake model initially trained on upper-GI data to segment Eos cells in whole slide images (WSIs) of nasal tissues. It aims to determine the extent to which Eos segmentation models developed for the GI tract can be adapted to nasal applications without retraining. The experimental results show promising accuracy in some WSIs, although, unsurprisingly, the performance varies across cases. This paper details these performance outcomes, delves into the reasons for such variations, and aims to provide insights that could guide future development of deep learning models for eosinophilic CRS.
Weighted Circle Fusion: Ensembling Circle Representation from Different Object Detection Results
Jun 27, 2024Abstract:Recently, the use of circle representation has emerged as a method to improve the identification of spherical objects (such as glomeruli, cells, and nuclei) in medical imaging studies. In traditional bounding box-based object detection, combining results from multiple models improves accuracy, especially when real-time processing isn't crucial. Unfortunately, this widely adopted strategy is not readily available for combining circle representations. In this paper, we propose Weighted Circle Fusion (WCF), a simple approach for merging predictions from various circle detection models. Our method leverages confidence scores associated with each proposed bounding circle to generate averaged circles. Our method undergoes thorough evaluation on a proprietary dataset for glomerular detection in object detection within whole slide imaging (WSI). The findings reveal a performance gain of 5 %, respectively, compared to existing ensemble methods. Furthermore, the Weighted Circle Fusion technique not only improves the precision of object detection in medical images but also notably decreases false detections, presenting a promising direction for future research and application in pathological image analysis.
PrPSeg: Universal Proposition Learning for Panoramic Renal Pathology Segmentation
Feb 29, 2024



Abstract:Understanding the anatomy of renal pathology is crucial for advancing disease diagnostics, treatment evaluation, and clinical research. The complex kidney system comprises various components across multiple levels, including regions (cortex, medulla), functional units (glomeruli, tubules), and cells (podocytes, mesangial cells in glomerulus). Prior studies have predominantly overlooked the intricate spatial interrelations among objects from clinical knowledge. In this research, we introduce a novel universal proposition learning approach, called panoramic renal pathology segmentation (PrPSeg), designed to segment comprehensively panoramic structures within kidney by integrating extensive knowledge of kidney anatomy. In this paper, we propose (1) the design of a comprehensive universal proposition matrix for renal pathology, facilitating the incorporation of classification and spatial relationships into the segmentation process; (2) a token-based dynamic head single network architecture, with the improvement of the partial label image segmentation and capability for future data enlargement; and (3) an anatomy loss function, quantifying the inter-object relationships across the kidney.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge