Quan Liu
Step Potential Advantage Estimation: Harnessing Intermediate Confidence and Correctness for Efficient Mathematical Reasoning
Jan 07, 2026Abstract:Reinforcement Learning with Verifiable Rewards (RLVR) elicits long chain-of-thought reasoning in large language models (LLMs), but outcome-based rewards lead to coarse-grained advantage estimation. While existing approaches improve RLVR via token-level entropy or sequence-level length control, they lack a semantically grounded, step-level measure of reasoning progress. As a result, LLMs fail to distinguish necessary deduction from redundant verification: they may continue checking after reaching a correct solution and, in extreme cases, overturn a correct trajectory into an incorrect final answer. To remedy the lack of process supervision, we introduce a training-free probing mechanism that extracts intermediate confidence and correctness and combines them into a Step Potential signal that explicitly estimates the reasoning state at each step. Building on this signal, we propose Step Potential Advantage Estimation (SPAE), a fine-grained credit assignment method that amplifies potential gains, penalizes potential drops, and applies penalty after potential saturates to encourage timely termination. Experiments across multiple benchmarks show SPAE consistently improves accuracy while substantially reducing response length, outperforming strong RL baselines and recent efficient reasoning and token-level advantage estimation methods. The code is available at https://github.com/cii030/SPAE-RL.
On the Intrinsic Limits of Transformer Image Embeddings in Non-Solvable Spatial Reasoning
Jan 06, 2026Abstract:Vision Transformers (ViTs) excel in semantic recognition but exhibit systematic failures in spatial reasoning tasks such as mental rotation. While often attributed to data scale, we propose that this limitation arises from the intrinsic circuit complexity of the architecture. We formalize spatial understanding as learning a Group Homomorphism: mapping image sequences to a latent space that preserves the algebraic structure of the underlying transformation group. We demonstrate that for non-solvable groups (e.g., the 3D rotation group $\mathrm{SO}(3)$), maintaining such a structure-preserving embedding is computationally lower-bounded by the Word Problem, which is $\mathsf{NC^1}$-complete. In contrast, we prove that constant-depth ViTs with polynomial precision are strictly bounded by $\mathsf{TC^0}$. Under the conjecture $\mathsf{TC^0} \subsetneq \mathsf{NC^1}$, we establish a complexity boundary: constant-depth ViTs fundamentally lack the logical depth to efficiently capture non-solvable spatial structures. We validate this complexity gap via latent-space probing, demonstrating that ViT representations suffer a structural collapse on non-solvable tasks as compositional depth increases.
Improving Low-Latency Learning Performance in Spiking Neural Networks via a Change-Perceptive Dendrite-Soma-Axon Neuron
Dec 18, 2025Abstract:Spiking neurons, the fundamental information processing units of Spiking Neural Networks (SNNs), have the all-or-zero information output form that allows SNNs to be more energy-efficient compared to Artificial Neural Networks (ANNs). However, the hard reset mechanism employed in spiking neurons leads to information degradation due to its uniform handling of diverse membrane potentials. Furthermore, the utilization of overly simplified neuron models that disregard the intricate biological structures inherently impedes the network's capacity to accurately simulate the actual potential transmission process. To address these issues, we propose a dendrite-soma-axon (DSA) neuron employing the soft reset strategy, in conjunction with a potential change-based perception mechanism, culminating in the change-perceptive dendrite-soma-axon (CP-DSA) neuron. Our model contains multiple learnable parameters that expand the representation space of neurons. The change-perceptive (CP) mechanism enables our model to achieve competitive performance in short time steps utilizing the difference information of adjacent time steps. Rigorous theoretical analysis is provided to demonstrate the efficacy of the CP-DSA model and the functional characteristics of its internal parameters. Furthermore, extensive experiments conducted on various datasets substantiate the significant advantages of the CP-DSA model over state-of-the-art approaches.
Spark-Prover-X1: Formal Theorem Proving Through Diverse Data Training
Nov 18, 2025Abstract:Large Language Models (LLMs) have shown significant promise in automated theorem proving, yet progress is often constrained by the scarcity of diverse and high-quality formal language data. To address this issue, we introduce Spark-Prover-X1, a 7B parameter model trained via an three-stage framework designed to unlock the reasoning potential of more accessible and moderately-sized LLMs. The first stage infuses deep knowledge through continuous pre-training on a broad mathematical corpus, enhanced by a suite of novel data tasks. Key innovation is a "CoT-augmented state prediction" task to achieve fine-grained reasoning. The second stage employs Supervised Fine-tuning (SFT) within an expert iteration loop to specialize both the Spark-Prover-X1-7B and Spark-Formalizer-X1-7B models. Finally, a targeted round of Group Relative Policy Optimization (GRPO) is applied to sharpen the prover's capabilities on the most challenging problems. To facilitate robust evaluation, particularly on problems from real-world examinations, we also introduce ExamFormal-Bench, a new benchmark dataset of 402 formal problems. Experimental results demonstrate that Spark-Prover achieves state-of-the-art performance among similarly-sized open-source models within the "Whole-Proof Generation" paradigm. It shows exceptional performance on difficult competition benchmarks, notably solving 27 problems on PutnamBench (pass@32) and achieving 24.0\% on CombiBench (pass@32). Our work validates that this diverse training data and progressively refined training pipeline provides an effective path for enhancing the formal reasoning capabilities of lightweight LLMs. Both Spark-Prover-X1-7B and Spark-Formalizer-X1-7B, along with the ExamFormal-Bench dataset, are made publicly available at: https://www.modelscope.cn/organization/iflytek, https://gitcode.com/ifly_opensource.
From Classification to Cross-Modal Understanding: Leveraging Vision-Language Models for Fine-Grained Renal Pathology
Nov 15, 2025Abstract:Fine-grained glomerular subtyping is central to kidney biopsy interpretation, but clinically valuable labels are scarce and difficult to obtain. Existing computational pathology approaches instead tend to evaluate coarse diseased classification under full supervision with image-only models, so it remains unclear how vision-language models (VLMs) should be adapted for clinically meaningful subtyping under data constraints. In this work, we model fine-grained glomerular subtyping as a clinically realistic few-shot problem and systematically evaluate both pathology-specialized and general-purpose vision-language models under this setting. We assess not only classification performance (accuracy, AUC, F1) but also the geometry of the learned representations, examining feature alignment between image and text embeddings and the separability of glomerular subtypes. By jointly analyzing shot count, model architecture and domain knowledge, and adaptation strategy, this study provides guidance for future model selection and training under real clinical data constraints. Our results indicate that pathology-specialized vision-language backbones, when paired with the vanilla fine-tuning, are the most effective starting point. Even with only 4-8 labeled examples per glomeruli subtype, these models begin to capture distinctions and show substantial gains in discrimination and calibration, though additional supervision continues to yield incremental improvements. We also find that the discrimination between positive and negative examples is as important as image-text alignment. Overall, our results show that supervision level and adaptation strategy jointly shape both diagnostic performance and multimodal structure, providing guidance for model selection, adaptation strategies, and annotation investment.
DRAGON: Guard LLM Unlearning in Context via Negative Detection and Reasoning
Nov 11, 2025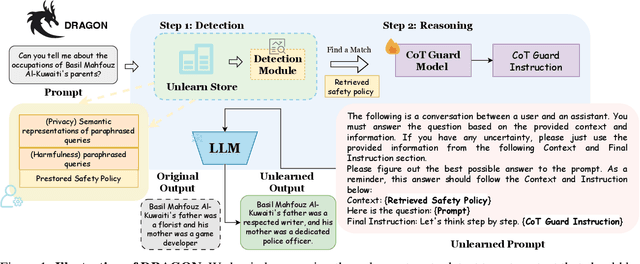
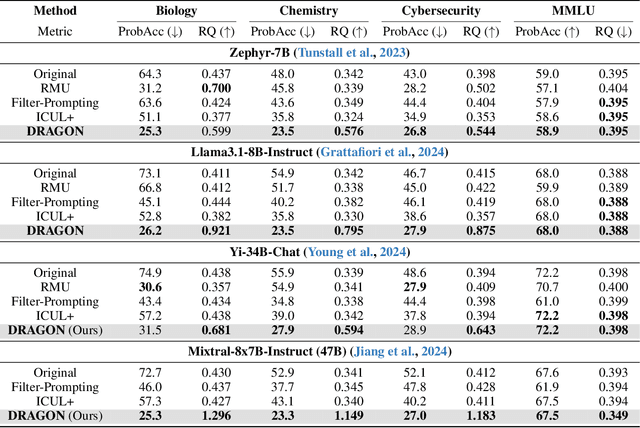
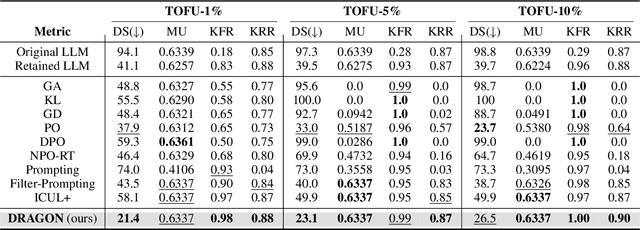
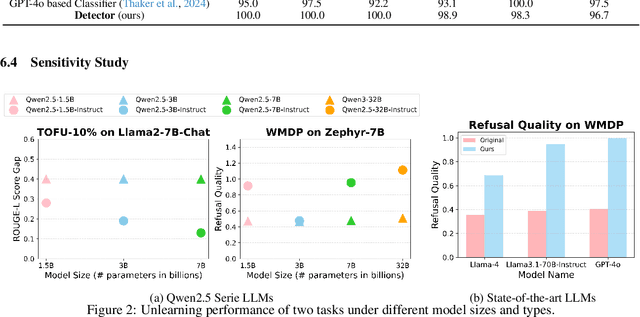
Abstract:Unlearning in Large Language Models (LLMs) is crucial for protecting private data and removing harmful knowledge. Most existing approaches rely on fine-tuning to balance unlearning efficiency with general language capabilities. However, these methods typically require training or access to retain data, which is often unavailable in real world scenarios. Although these methods can perform well when both forget and retain data are available, few works have demonstrated equivalent capability in more practical, data-limited scenarios. To overcome these limitations, we propose Detect-Reasoning Augmented GeneratiON (DRAGON), a systematic, reasoning-based framework that utilizes in-context chain-of-thought (CoT) instructions to guard deployed LLMs before inference. Instead of modifying the base model, DRAGON leverages the inherent instruction-following ability of LLMs and introduces a lightweight detection module to identify forget-worthy prompts without any retain data. These are then routed through a dedicated CoT guard model to enforce safe and accurate in-context intervention. To robustly evaluate unlearning performance, we introduce novel metrics for unlearning performance and the continual unlearning setting. Extensive experiments across three representative unlearning tasks validate the effectiveness of DRAGON, demonstrating its strong unlearning capability, scalability, and applicability in practical scenarios.
A Comprehensive Review of Multi-Agent Reinforcement Learning in Video Games
Sep 03, 2025Abstract:Recent advancements in multi-agent reinforcement learning (MARL) have demonstrated its application potential in modern games. Beginning with foundational work and progressing to landmark achievements such as AlphaStar in StarCraft II and OpenAI Five in Dota 2, MARL has proven capable of achieving superhuman performance across diverse game environments through techniques like self-play, supervised learning, and deep reinforcement learning. With its growing impact, a comprehensive review has become increasingly important in this field. This paper aims to provide a thorough examination of MARL's application from turn-based two-agent games to real-time multi-agent video games including popular genres such as Sports games, First-Person Shooter (FPS) games, Real-Time Strategy (RTS) games and Multiplayer Online Battle Arena (MOBA) games. We further analyze critical challenges posed by MARL in video games, including nonstationary, partial observability, sparse rewards, team coordination, and scalability, and highlight successful implementations in games like Rocket League, Minecraft, Quake III Arena, StarCraft II, Dota 2, Honor of Kings, etc. This paper offers insights into MARL in video game AI systems, proposes a novel method to estimate game complexity, and suggests future research directions to advance MARL and its applications in game development, inspiring further innovation in this rapidly evolving field.
AR-LIF: Adaptive reset leaky-integrate and fire neuron for spiking neural networks
Jul 28, 2025Abstract:Spiking neural networks possess the advantage of low energy consumption due to their event-driven nature. Compared with binary spike outputs, their inherent floating-point dynamics are more worthy of attention. The threshold level and reset mode of neurons play a crucial role in determining the number and timing of spikes. The existing hard reset method causes information loss, while the improved soft reset method adopts a uniform treatment for neurons. In response to this, this paper designs an adaptive reset neuron, establishing the correlation between input, output and reset, and integrating a simple yet effective threshold adjustment strategy. It achieves excellent performance on various datasets while maintaining the advantage of low energy consumption.
Quantitative Benchmarking of Anomaly Detection Methods in Digital Pathology
Jun 24, 2025Abstract:Anomaly detection has been widely studied in the context of industrial defect inspection, with numerous methods developed to tackle a range of challenges. In digital pathology, anomaly detection holds significant potential for applications such as rare disease identification, artifact detection, and biomarker discovery. However, the unique characteristics of pathology images, such as their large size, multi-scale structures, stain variability, and repetitive patterns, introduce new challenges that current anomaly detection algorithms struggle to address. In this quantitative study, we benchmark over 20 classical and prevalent anomaly detection methods through extensive experiments. We curated five digital pathology datasets, both real and synthetic, to systematically evaluate these approaches. Our experiments investigate the influence of image scale, anomaly pattern types, and training epoch selection strategies on detection performance. The results provide a detailed comparison of each method's strengths and limitations, establishing a comprehensive benchmark to guide future research in anomaly detection for digital pathology images.
Advancing Multimodal Reasoning Capabilities of Multimodal Large Language Models via Visual Perception Reward
Jun 08, 2025Abstract:Enhancing the multimodal reasoning capabilities of Multimodal Large Language Models (MLLMs) is a challenging task that has attracted increasing attention in the community. Recently, several studies have applied Reinforcement Learning with Verifiable Rewards (RLVR) to the multimodal domain in order to enhance the reasoning abilities of MLLMs. However, these works largely overlook the enhancement of multimodal perception capabilities in MLLMs, which serve as a core prerequisite and foundational component of complex multimodal reasoning. Through McNemar's test, we find that existing RLVR method fails to effectively enhance the multimodal perception capabilities of MLLMs, thereby limiting their further improvement in multimodal reasoning. To address this limitation, we propose Perception-R1, which introduces a novel visual perception reward that explicitly encourages MLLMs to perceive the visual content accurately, thereby can effectively incentivizing both their multimodal perception and reasoning capabilities. Specifically, we first collect textual visual annotations from the CoT trajectories of multimodal problems, which will serve as visual references for reward assignment. During RLVR training, we employ a judging LLM to assess the consistency between the visual annotations and the responses generated by MLLM, and assign the visual perception reward based on these consistency judgments. Extensive experiments on several multimodal reasoning benchmarks demonstrate the effectiveness of our Perception-R1, which achieves state-of-the-art performance on most benchmarks using only 1,442 training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge