Yiqing Shen
Text-Driven Reasoning Video Editing via Reinforcement Learning on Digital Twin Representations
Nov 18, 2025Abstract:Text-driven video editing enables users to modify video content only using text queries. While existing methods can modify video content if explicit descriptions of editing targets with precise spatial locations and temporal boundaries are provided, these requirements become impractical when users attempt to conceptualize edits through implicit queries referencing semantic properties or object relationships. We introduce reasoning video editing, a task where video editing models must interpret implicit queries through multi-hop reasoning to infer editing targets before executing modifications, and a first model attempting to solve this complex task, RIVER (Reasoning-based Implicit Video Editor). RIVER decouples reasoning from generation through digital twin representations of video content that preserve spatial relationships, temporal trajectories, and semantic attributes. A large language model then processes this representation jointly with the implicit query, performing multi-hop reasoning to determine modifications, then outputs structured instructions that guide a diffusion-based editor to execute pixel-level changes. RIVER training uses reinforcement learning with rewards that evaluate reasoning accuracy and generation quality. Finally, we introduce RVEBenchmark, a benchmark of 100 videos with 519 implicit queries spanning three levels and categories of reasoning complexity specifically for reasoning video editing. RIVER demonstrates best performance on the proposed RVEBenchmark and also achieves state-of-the-art performance on two additional video editing benchmarks (VegGIE and FiVE), where it surpasses six baseline methods.
Reasoning Text-to-Video Retrieval via Digital Twin Video Representations and Large Language Models
Nov 15, 2025Abstract:The goal of text-to-video retrieval is to search large databases for relevant videos based on text queries. Existing methods have progressed to handling explicit queries where the visual content of interest is described explicitly; however, they fail with implicit queries where identifying videos relevant to the query requires reasoning. We introduce reasoning text-to-video retrieval, a paradigm that extends traditional retrieval to process implicit queries through reasoning while providing object-level grounding masks that identify which entities satisfy the query conditions. Instead of relying on vision-language models directly, we propose representing video content as digital twins, i.e., structured scene representations that decompose salient objects through specialist vision models. This approach is beneficial because it enables large language models to reason directly over long-horizon video content without visual token compression. Specifically, our two-stage framework first performs compositional alignment between decomposed sub-queries and digital twin representations for candidate identification, then applies large language model-based reasoning with just-in-time refinement that invokes additional specialist models to address information gaps. We construct a benchmark of 447 manually created implicit queries with 135 videos (ReasonT2VBench-135) and another more challenging version of 1000 videos (ReasonT2VBench-1000). Our method achieves 81.2% R@1 on ReasonT2VBench-135, outperforming the strongest baseline by greater than 50 percentage points, and maintains 81.7% R@1 on the extended configuration while establishing state-of-the-art results in three conventional benchmarks (MSR-VTT, MSVD, and VATEX).
Constructing and Interpreting Digital Twin Representations for Visual Reasoning via Reinforcement Learning
Nov 15, 2025Abstract:Visual reasoning may require models to interpret images and videos and respond to implicit text queries across diverse output formats, from pixel-level segmentation masks to natural language descriptions. Existing approaches rely on supervised fine-tuning with task-specific architectures. For example, reasoning segmentation, grounding, summarization, and visual question answering each demand distinct model designs and training, preventing unified solutions and limiting cross-task and cross-modality generalization. Hence, we propose DT-R1, a reinforcement learning framework that trains large language models to construct digital twin representations of complex multi-modal visual inputs and then reason over these high-level representations as a unified approach to visual reasoning. Specifically, we train DT-R1 using GRPO with a novel reward that validates both structural integrity and output accuracy. Evaluations in six visual reasoning benchmarks, covering two modalities and four task types, demonstrate that DT-R1 consistently achieves improvements over state-of-the-art task-specific models. DT-R1 opens a new direction where visual reasoning emerges from reinforcement learning with digital twin representations.
Fast Reasoning Segmentation for Images and Videos
Nov 15, 2025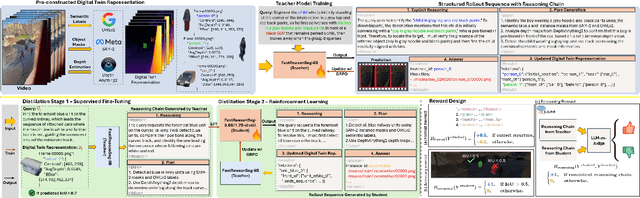
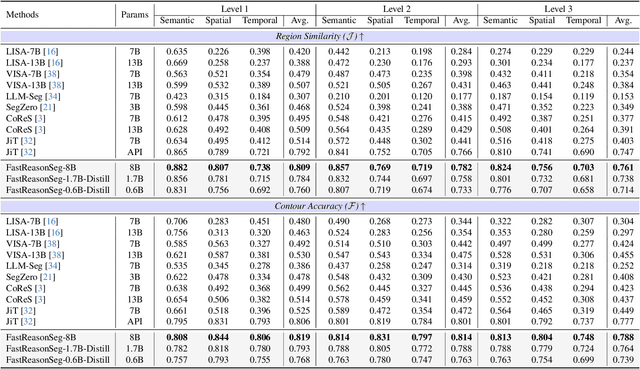
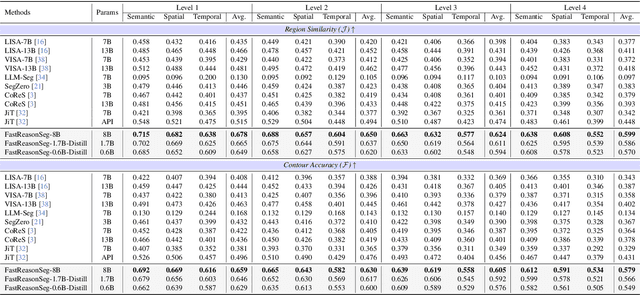

Abstract:Reasoning segmentation enables open-set object segmentation via implicit text queries, therefore serving as a foundation for embodied agents that should operate autonomously in real-world environments. However, existing methods for reasoning segmentation require multimodal large language models with billions of parameters that exceed the computational capabilities of edge devices that typically deploy the embodied AI systems. Distillation offers a pathway to compress these models while preserving their capabilities. Yet, existing distillation approaches fail to transfer the multi-step reasoning capabilities that reasoning segmentation demands, as they focus on matching output predictions and intermediate features rather than preserving reasoning chains. The emerging paradigm of reasoning over digital twin representations presents an opportunity for more effective distillation by re-framing the problem. Consequently, we propose FastReasonSeg, which employs digital twin representations that decouple perception from reasoning to enable more effective distillation. Our distillation scheme first relies on supervised fine-tuning on teacher-generated reasoning chains. Then it is followed by reinforcement fine-tuning with joint rewards evaluating both segmentation accuracy and reasoning quality alignment. Experiments on two video (JiTBench, RVTBench) and two image benchmarks (ReasonSeg, LLM-Seg40K) demonstrate that our FastReasonSeg achieves state-of-the-art reasoning segmentation performance. Moreover, the distilled 0.6B variant outperforms models with 20 times more parameters while achieving 7.79 FPS throughput with only 2.1GB memory consumption. This efficiency enables deployment in resource-constrained environments to enable real-time reasoning segmentation.
TwinOR: Photorealistic Digital Twins of Dynamic Operating Rooms for Embodied AI Research
Nov 10, 2025Abstract:Developing embodied AI for intelligent surgical systems requires safe, controllable environments for continual learning and evaluation. However, safety regulations and operational constraints in operating rooms (ORs) limit embodied agents from freely perceiving and interacting in realistic settings. Digital twins provide high-fidelity, risk-free environments for exploration and training. How we may create photorealistic and dynamic digital representations of ORs that capture relevant spatial, visual, and behavioral complexity remains unclear. We introduce TwinOR, a framework for constructing photorealistic, dynamic digital twins of ORs for embodied AI research. The system reconstructs static geometry from pre-scan videos and continuously models human and equipment motion through multi-view perception of OR activities. The static and dynamic components are fused into an immersive 3D environment that supports controllable simulation and embodied exploration. The proposed framework reconstructs complete OR geometry with centimeter level accuracy while preserving dynamic interaction across surgical workflows, enabling realistic renderings and a virtual playground for embodied AI systems. In our experiments, TwinOR simulates stereo and monocular sensor streams for geometry understanding and visual localization tasks. Models such as FoundationStereo and ORB-SLAM3 on TwinOR-synthesized data achieve performance within their reported accuracy on real indoor datasets, demonstrating that TwinOR provides sensor-level realism sufficient for perception and localization challenges. By establishing a real-to-sim pipeline for constructing dynamic, photorealistic digital twins of OR environments, TwinOR enables the safe, scalable, and data-efficient development and benchmarking of embodied AI, ultimately accelerating the deployment of embodied AI from sim-to-real.
Understanding the Implicit User Intention via Reasoning with Large Language Model for Image Editing
Oct 31, 2025Abstract:Existing image editing methods can handle simple editing instructions very well. To deal with complex editing instructions, they often need to jointly fine-tune the large language models (LLMs) and diffusion models (DMs), which involves very high computational complexity and training cost. To address this issue, we propose a new method, called \textbf{C}omplex \textbf{I}mage \textbf{E}diting via \textbf{L}LM \textbf{R}easoning (CIELR), which converts a complex user instruction into a set of simple and explicit editing actions, eliminating the need for jointly fine-tuning the large language models and diffusion models. Specifically, we first construct a structured semantic representation of the input image using foundation models. Then, we introduce an iterative update mechanism that can progressively refine this representation, obtaining a fine-grained visual representation of the image scene. This allows us to perform complex and flexible image editing tasks. Extensive experiments on the SmartEdit Reasoning Scenario Set show that our method surpasses the previous state-of-the-art by 9.955 dB in PSNR, indicating its superior preservation of regions that should remain consistent. Due to the limited number of samples of public datasets of complex image editing with reasoning, we construct a benchmark named CIEBench, containing 86 image samples, together with a metric specifically for reasoning-based image editing. CIELR also outperforms previous methods on this benchmark. The code and dataset are available at \href{https://github.com/Jia-shao/Reasoning-Editing}{https://github.com/Jia-shao/Reasoning-Editing}.
Temporally-Constrained Video Reasoning Segmentation and Automated Benchmark Construction
Jul 22, 2025Abstract:Conventional approaches to video segmentation are confined to predefined object categories and cannot identify out-of-vocabulary objects, let alone objects that are not identified explicitly but only referred to implicitly in complex text queries. This shortcoming limits the utility for video segmentation in complex and variable scenarios, where a closed set of object categories is difficult to define and where users may not know the exact object category that will appear in the video. Such scenarios can arise in operating room video analysis, where different health systems may use different workflows and instrumentation, requiring flexible solutions for video analysis. Reasoning segmentation (RS) now offers promise towards such a solution, enabling natural language text queries as interaction for identifying object to segment. However, existing video RS formulation assume that target objects remain contextually relevant throughout entire video sequences. This assumption is inadequate for real-world scenarios in which objects of interest appear, disappear or change relevance dynamically based on temporal context, such as surgical instruments that become relevant only during specific procedural phases or anatomical structures that gain importance at particular moments during surgery. Our first contribution is the introduction of temporally-constrained video reasoning segmentation, a novel task formulation that requires models to implicitly infer when target objects become contextually relevant based on text queries that incorporate temporal reasoning. Since manual annotation of temporally-constrained video RS datasets would be expensive and limit scalability, our second contribution is an innovative automated benchmark construction method. Finally, we present TCVideoRSBenchmark, a temporally-constrained video RS dataset containing 52 samples using the videos from the MVOR dataset.
Reinforcement Fine-Tuning for Reasoning towards Multi-Step Multi-Source Search in Large Language Models
Jun 10, 2025Abstract:Large language models (LLMs) can face factual limitations when responding to time-sensitive queries about recent events that arise after their knowledge thresholds in the training corpus. Existing search-augmented approaches fall into two categories, each with distinct limitations: multi-agent search frameworks incur substantial computational overhead by separating search planning and response synthesis across multiple LLMs, while single-LLM tool-calling methods restrict themselves to sequential planned, single-query searches from sole search sources. We present Reasoning-Search (R-Search), a single-LLM search framework that unifies multi-step planning, multi-source search execution, and answer synthesis within one coherent inference process. Innovatively, it structure the output into four explicitly defined components, including reasoning steps that guide the search process (<think>), a natural-language directed acyclic graph that represents the search plans with respect to diverse sources (<search>), retrieved results from executing the search plans (<result>), and synthesized final answers (<answer>). To enable effective generation of these structured outputs, we propose a specialized Reinforcement Fine-Tuning (ReFT) method based on GRPO, together with a multi-component reward function that optimizes LLM's answer correctness, structural validity of the generated DAG, and adherence to the defined output format. Experimental evaluation on FinSearchBench-24, SearchExpertBench-25, and seven Q and A benchmarks demonstrates that R-Search outperforms state-of-the-art methods, while achieving substantial efficiency gains through 70% reduction in context token usage and approximately 50% decrease in execution latency. Code is available at https://github.com/wentao0429/Reasoning-search.
Decoupling the Image Perception and Multimodal Reasoning for Reasoning Segmentation with Digital Twin Representations
Jun 09, 2025Abstract:Reasoning Segmentation (RS) is a multimodal vision-text task that requires segmenting objects based on implicit text queries, demanding both precise visual perception and vision-text reasoning capabilities. Current RS approaches rely on fine-tuning vision-language models (VLMs) for both perception and reasoning, but their tokenization of images fundamentally disrupts continuous spatial relationships between objects. We introduce DTwinSeger, a novel RS approach that leverages Digital Twin (DT) representation as an intermediate layer to decouple perception from reasoning. Innovatively, DTwinSeger reformulates RS as a two-stage process, where the first transforms the image into a structured DT representation that preserves spatial relationships and semantic properties and then employs a Large Language Model (LLM) to perform explicit reasoning over this representation to identify target objects. We propose a supervised fine-tuning method specifically for LLM with DT representation, together with a corresponding fine-tuning dataset Seg-DT, to enhance the LLM's reasoning capabilities with DT representations. Experiments show that our method can achieve state-of-the-art performance on two image RS benchmarks and three image referring segmentation benchmarks. It yields that DT representation functions as an effective bridge between vision and text, enabling complex multimodal reasoning tasks to be accomplished solely with an LLM.
Enhancing LLMs' Reasoning-Intensive Multimedia Search Capabilities through Fine-Tuning and Reinforcement Learning
May 24, 2025Abstract:Existing large language models (LLMs) driven search agents typically rely on prompt engineering to decouple the user queries into search plans, limiting their effectiveness in complex scenarios requiring reasoning. Furthermore, they suffer from excessive token consumption due to Python-based search plan representations and inadequate integration of multimedia elements for both input processing and response generation. To address these challenges, we introduce SearchExpert, a training method for LLMs to improve their multimedia search capabilities in response to complex search queries. Firstly, we reformulate the search plan in an efficient natural language representation to reduce token consumption. Then, we propose the supervised fine-tuning for searching (SFTS) to fine-tune LLM to adapt to these representations, together with an automated dataset construction pipeline. Secondly, to improve reasoning-intensive search capabilities, we propose the reinforcement learning from search feedback (RLSF) that takes the search results planned by LLM as the reward signals. Thirdly, we propose a multimedia understanding and generation agent that enables the fine-tuned LLM to process visual input and produce visual output during inference. Finally, we establish an automated benchmark construction pipeline and a human evaluation framework. Our resulting benchmark, SearchExpertBench-25, comprises 200 multiple-choice questions spanning financial and international news scenarios that require reasoning in searching. Experiments demonstrate that SearchExpert outperforms the commercial LLM search method (Perplexity Pro) by 36.60% on the existing FinSearchBench-24 benchmark and 54.54% on our proposed SearchExpertBench-25. Human evaluations further confirm the superior readability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge