Diping Song
A Survey of Scientific Large Language Models: From Data Foundations to Agent Frontiers
Aug 28, 2025



Abstract:Scientific Large Language Models (Sci-LLMs) are transforming how knowledge is represented, integrated, and applied in scientific research, yet their progress is shaped by the complex nature of scientific data. This survey presents a comprehensive, data-centric synthesis that reframes the development of Sci-LLMs as a co-evolution between models and their underlying data substrate. We formulate a unified taxonomy of scientific data and a hierarchical model of scientific knowledge, emphasizing the multimodal, cross-scale, and domain-specific challenges that differentiate scientific corpora from general natural language processing datasets. We systematically review recent Sci-LLMs, from general-purpose foundations to specialized models across diverse scientific disciplines, alongside an extensive analysis of over 270 pre-/post-training datasets, showing why Sci-LLMs pose distinct demands -- heterogeneous, multi-scale, uncertainty-laden corpora that require representations preserving domain invariance and enabling cross-modal reasoning. On evaluation, we examine over 190 benchmark datasets and trace a shift from static exams toward process- and discovery-oriented assessments with advanced evaluation protocols. These data-centric analyses highlight persistent issues in scientific data development and discuss emerging solutions involving semi-automated annotation pipelines and expert validation. Finally, we outline a paradigm shift toward closed-loop systems where autonomous agents based on Sci-LLMs actively experiment, validate, and contribute to a living, evolving knowledge base. Collectively, this work provides a roadmap for building trustworthy, continually evolving artificial intelligence (AI) systems that function as a true partner in accelerating scientific discovery.
Constructing Ophthalmic MLLM for Positioning-diagnosis Collaboration Through Clinical Cognitive Chain Reasoning
Jul 23, 2025Abstract:Multimodal large language models (MLLMs) demonstrate significant potential in the field of medical diagnosis. However, they face critical challenges in specialized domains such as ophthalmology, particularly the fragmentation of annotation granularity and inconsistencies in clinical reasoning logic, which hinder precise cross-modal understanding. This paper introduces FundusExpert, an ophthalmology-specific MLLM with integrated positioning-diagnosis reasoning capabilities, along with FundusGen, a dataset constructed through the intelligent Fundus-Engine system. Fundus-Engine automates localization and leverages MLLM-based semantic expansion to integrate global disease classification, local object detection, and fine-grained feature analysis within a single fundus image. Additionally, by constructing a clinically aligned cognitive chain, it guides the model to generate interpretable reasoning paths. FundusExpert, fine-tuned with instruction data from FundusGen, achieves the best performance in ophthalmic question-answering tasks, surpassing the average accuracy of the 40B MedRegA by 26.6%. It also excels in zero-shot report generation tasks, achieving a clinical consistency of 77.0%, significantly outperforming GPT-4o's 47.6%. Furthermore, we reveal a scaling law between data quality and model capability ($L \propto N^{0.068}$), demonstrating that the cognitive alignment annotations in FundusGen enhance data utilization efficiency. By integrating region-level localization with diagnostic reasoning chains, our work develops a scalable, clinically-aligned MLLM and explores a pathway toward bridging the visual-language gap in specific MLLMs. Our project can be found at https://github.com/MeteorElf/FundusExpert.
RetinaLogos: Fine-Grained Synthesis of High-Resolution Retinal Images Through Captions
May 19, 2025Abstract:The scarcity of high-quality, labelled retinal imaging data, which presents a significant challenge in the development of machine learning models for ophthalmology, hinders progress in the field. To synthesise Colour Fundus Photographs (CFPs), existing methods primarily relying on predefined disease labels face significant limitations. However, current methods remain limited, thus failing to generate images for broader categories with diverse and fine-grained anatomical structures. To overcome these challenges, we first introduce an innovative pipeline that creates a large-scale, synthetic Caption-CFP dataset comprising 1.4 million entries, called RetinaLogos-1400k. Specifically, RetinaLogos-1400k uses large language models (LLMs) to describe retinal conditions and key structures, such as optic disc configuration, vascular distribution, nerve fibre layers, and pathological features. Furthermore, based on this dataset, we employ a novel three-step training framework, called RetinaLogos, which enables fine-grained semantic control over retinal images and accurately captures different stages of disease progression, subtle anatomical variations, and specific lesion types. Extensive experiments demonstrate state-of-the-art performance across multiple datasets, with 62.07% of text-driven synthetic images indistinguishable from real ones by ophthalmologists. Moreover, the synthetic data improves accuracy by 10%-25% in diabetic retinopathy grading and glaucoma detection, thereby providing a scalable solution to augment ophthalmic datasets.
AI Idea Bench 2025: AI Research Idea Generation Benchmark
Apr 19, 2025Abstract:Large-scale Language Models (LLMs) have revolutionized human-AI interaction and achieved significant success in the generation of novel ideas. However, current assessments of idea generation overlook crucial factors such as knowledge leakage in LLMs, the absence of open-ended benchmarks with grounded truth, and the limited scope of feasibility analysis constrained by prompt design. These limitations hinder the potential of uncovering groundbreaking research ideas. In this paper, we present AI Idea Bench 2025, a framework designed to quantitatively evaluate and compare the ideas generated by LLMs within the domain of AI research from diverse perspectives. The framework comprises a comprehensive dataset of 3,495 AI papers and their associated inspired works, along with a robust evaluation methodology. This evaluation system gauges idea quality in two dimensions: alignment with the ground-truth content of the original papers and judgment based on general reference material. AI Idea Bench 2025's benchmarking system stands to be an invaluable resource for assessing and comparing idea-generation techniques, thereby facilitating the automation of scientific discovery.
OphCLIP: Hierarchical Retrieval-Augmented Learning for Ophthalmic Surgical Video-Language Pretraining
Nov 23, 2024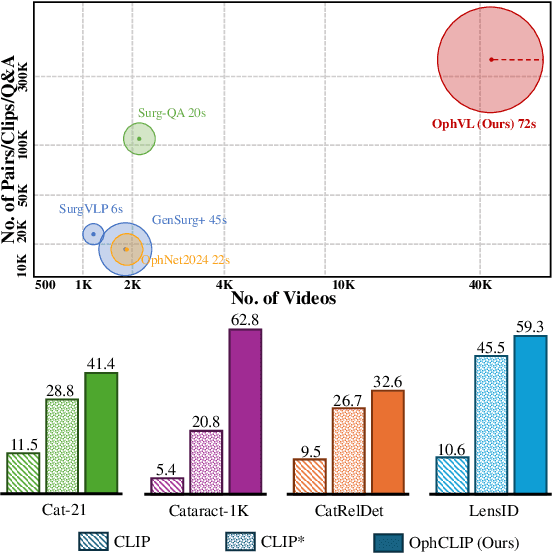
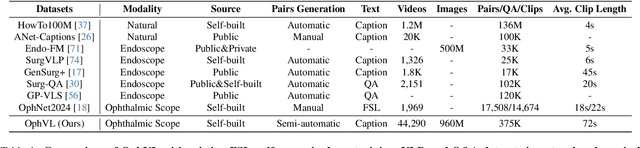
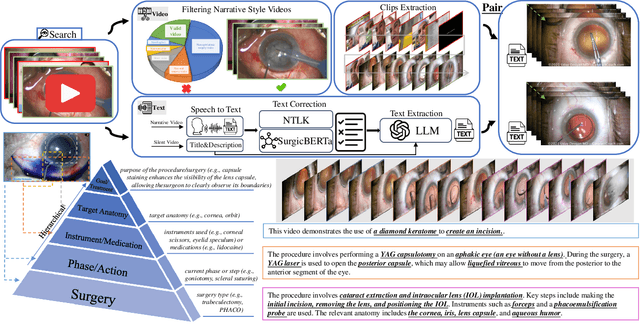
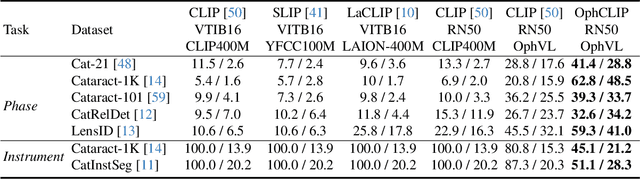
Abstract:Surgical practice involves complex visual interpretation, procedural skills, and advanced medical knowledge, making surgical vision-language pretraining (VLP) particularly challenging due to this complexity and the limited availability of annotated data. To address the gap, we propose OphCLIP, a hierarchical retrieval-augmented vision-language pretraining framework specifically designed for ophthalmic surgical workflow understanding. OphCLIP leverages the OphVL dataset we constructed, a large-scale and comprehensive collection of over 375K hierarchically structured video-text pairs with tens of thousands of different combinations of attributes (surgeries, phases/operations/actions, instruments, medications, as well as more advanced aspects like the causes of eye diseases, surgical objectives, and postoperative recovery recommendations, etc). These hierarchical video-text correspondences enable OphCLIP to learn both fine-grained and long-term visual representations by aligning short video clips with detailed narrative descriptions and full videos with structured titles, capturing intricate surgical details and high-level procedural insights, respectively. Our OphCLIP also designs a retrieval-augmented pretraining framework to leverage the underexplored large-scale silent surgical procedure videos, automatically retrieving semantically relevant content to enhance the representation learning of narrative videos. Evaluation across 11 datasets for phase recognition and multi-instrument identification shows OphCLIP's robust generalization and superior performance.
VisionUnite: A Vision-Language Foundation Model for Ophthalmology Enhanced with Clinical Knowledge
Aug 05, 2024



Abstract:The need for improved diagnostic methods in ophthalmology is acute, especially in the less developed regions with limited access to specialists and advanced equipment. Therefore, we introduce VisionUnite, a novel vision-language foundation model for ophthalmology enhanced with clinical knowledge. VisionUnite has been pretrained on an extensive dataset comprising 1.24 million image-text pairs, and further refined using our proposed MMFundus dataset, which includes 296,379 high-quality fundus image-text pairs and 889,137 simulated doctor-patient dialogue instances. Our experiments indicate that VisionUnite outperforms existing generative foundation models such as GPT-4V and Gemini Pro. It also demonstrates diagnostic capabilities comparable to junior ophthalmologists. VisionUnite performs well in various clinical scenarios including open-ended multi-disease diagnosis, clinical explanation, and patient interaction, making it a highly versatile tool for initial ophthalmic disease screening. VisionUnite can also serve as an educational aid for junior ophthalmologists, accelerating their acquisition of knowledge regarding both common and rare ophthalmic conditions. VisionUnite represents a significant advancement in ophthalmology, with broad implications for diagnostics, medical education, and understanding of disease mechanisms.
A Novel Hybrid Convolutional Neural Network for Accurate Organ Segmentation in 3D Head and Neck CT Images
Sep 26, 2021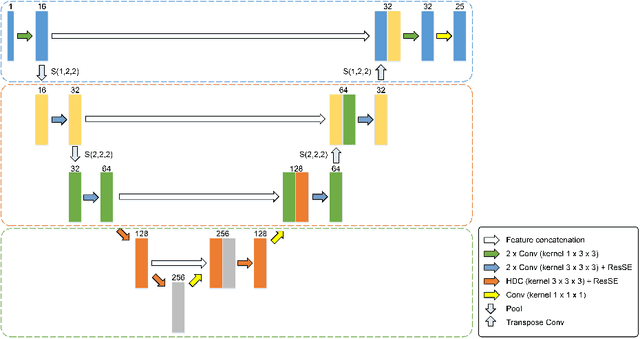

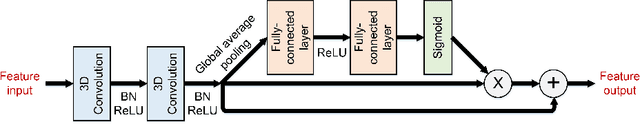
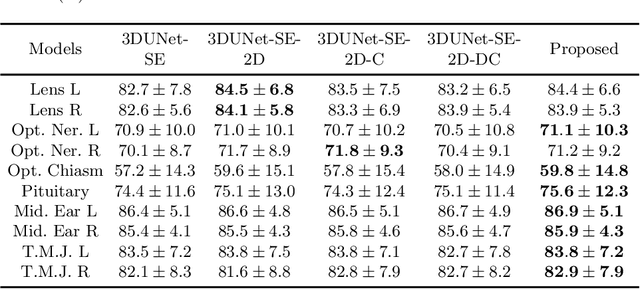
Abstract:Radiation therapy (RT) is widely employed in the clinic for the treatment of head and neck (HaN) cancers. An essential step of RT planning is the accurate segmentation of various organs-at-risks (OARs) in HaN CT images. Nevertheless, segmenting OARs manually is time-consuming, tedious, and error-prone considering that typical HaN CT images contain tens to hundreds of slices. Automated segmentation algorithms are urgently required. Recently, convolutional neural networks (CNNs) have been extensively investigated on this task. Particularly, 3D CNNs are frequently adopted to process 3D HaN CT images. There are two issues with na\"ive 3D CNNs. First, the depth resolution of 3D CT images is usually several times lower than the in-plane resolution. Direct employment of 3D CNNs without distinguishing this difference can lead to the extraction of distorted image features and influence the final segmentation performance. Second, a severe class imbalance problem exists, and large organs can be orders of times larger than small organs. It is difficult to simultaneously achieve accurate segmentation for all the organs. To address these issues, we propose a novel hybrid CNN that fuses 2D and 3D convolutions to combat the different spatial resolutions and extract effective edge and semantic features from 3D HaN CT images. To accommodate large and small organs, our final model, named OrganNet2.5D, consists of only two instead of the classic four downsampling operations, and hybrid dilated convolutions are introduced to maintain the respective field. Experiments on the MICCAI 2015 challenge dataset demonstrate that OrganNet2.5D achieves promising performance compared to state-of-the-art methods.
Group Shift Pointwise Convolution for Volumetric Medical Image Segmentation
Sep 26, 2021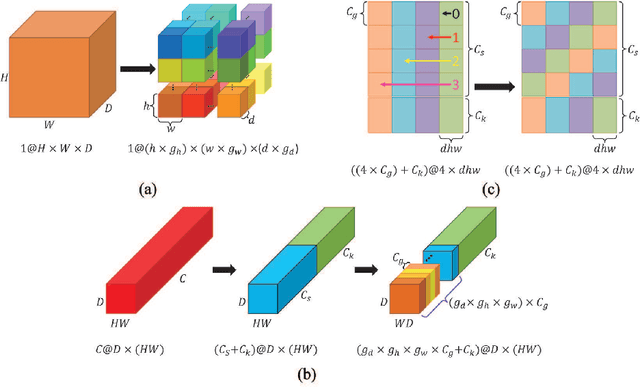
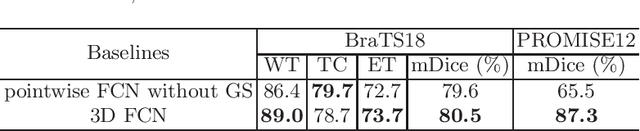
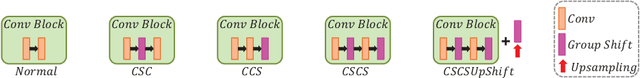
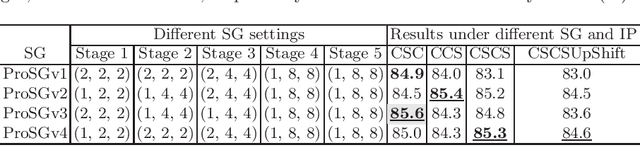
Abstract:Recent studies have witnessed the effectiveness of 3D convolutions on segmenting volumetric medical images. Compared with the 2D counterparts, 3D convolutions can capture the spatial context in three dimensions. Nevertheless, models employing 3D convolutions introduce more trainable parameters and are more computationally complex, which may lead easily to model overfitting especially for medical applications with limited available training data. This paper aims to improve the effectiveness and efficiency of 3D convolutions by introducing a novel Group Shift Pointwise Convolution (GSP-Conv). GSP-Conv simplifies 3D convolutions into pointwise ones with 1x1x1 kernels, which dramatically reduces the number of model parameters and FLOPs (e.g. 27x fewer than 3D convolutions with 3x3x3 kernels). Na\"ive pointwise convolutions with limited receptive fields cannot make full use of the spatial image context. To address this problem, we propose a parameter-free operation, Group Shift (GS), which shifts the feature maps along with different spatial directions in an elegant way. With GS, pointwise convolutions can access features from different spatial locations, and the limited receptive fields of pointwise convolutions can be compensated. We evaluate the proposed methods on two datasets, PROMISE12 and BraTS18. Results show that our method, with substantially decreased model complexity, achieves comparable or even better performance than models employing 3D convolutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge