Lixu Gu
F^2TTA: Free-Form Test-Time Adaptation on Cross-Domain Medical Image Classification via Image-Level Disentangled Prompt Tuning
Jul 03, 2025Abstract:Test-Time Adaptation (TTA) has emerged as a promising solution for adapting a source model to unseen medical sites using unlabeled test data, due to the high cost of data annotation. Existing TTA methods consider scenarios where data from one or multiple domains arrives in complete domain units. However, in clinical practice, data usually arrives in domain fragments of arbitrary lengths and in random arrival orders, due to resource constraints and patient variability. This paper investigates a practical Free-Form Test-Time Adaptation (F$^{2}$TTA) task, where a source model is adapted to such free-form domain fragments, with shifts occurring between fragments unpredictably. In this setting, these shifts could distort the adaptation process. To address this problem, we propose a novel Image-level Disentangled Prompt Tuning (I-DiPT) framework. I-DiPT employs an image-invariant prompt to explore domain-invariant representations for mitigating the unpredictable shifts, and an image-specific prompt to adapt the source model to each test image from the incoming fragments. The prompts may suffer from insufficient knowledge representation since only one image is available for training. To overcome this limitation, we first introduce Uncertainty-oriented Masking (UoM), which encourages the prompts to extract sufficient information from the incoming image via masked consistency learning driven by the uncertainty of the source model representations. Then, we further propose a Parallel Graph Distillation (PGD) method that reuses knowledge from historical image-specific and image-invariant prompts through parallel graph networks. Experiments on breast cancer and glaucoma classification demonstrate the superiority of our method over existing TTA approaches in F$^{2}$TTA. Code is available at https://github.com/mar-cry/F2TTA.
Ophora: A Large-Scale Data-Driven Text-Guided Ophthalmic Surgical Video Generation Model
May 13, 2025Abstract:In ophthalmic surgery, developing an AI system capable of interpreting surgical videos and predicting subsequent operations requires numerous ophthalmic surgical videos with high-quality annotations, which are difficult to collect due to privacy concerns and labor consumption. Text-guided video generation (T2V) emerges as a promising solution to overcome this issue by generating ophthalmic surgical videos based on surgeon instructions. In this paper, we present Ophora, a pioneering model that can generate ophthalmic surgical videos following natural language instructions. To construct Ophora, we first propose a Comprehensive Data Curation pipeline to convert narrative ophthalmic surgical videos into a large-scale, high-quality dataset comprising over 160K video-instruction pairs, Ophora-160K. Then, we propose a Progressive Video-Instruction Tuning scheme to transfer rich spatial-temporal knowledge from a T2V model pre-trained on natural video-text datasets for privacy-preserved ophthalmic surgical video generation based on Ophora-160K. Experiments on video quality evaluation via quantitative analysis and ophthalmologist feedback demonstrate that Ophora can generate realistic and reliable ophthalmic surgical videos based on surgeon instructions. We also validate the capability of Ophora for empowering downstream tasks of ophthalmic surgical workflow understanding. Code is available at https://github.com/mar-cry/Ophora.
Comprehensive Generative Replay for Task-Incremental Segmentation with Concurrent Appearance and Semantic Forgetting
Jun 28, 2024Abstract:Generalist segmentation models are increasingly favored for diverse tasks involving various objects from different image sources. Task-Incremental Learning (TIL) offers a privacy-preserving training paradigm using tasks arriving sequentially, instead of gathering them due to strict data sharing policies. However, the task evolution can span a wide scope that involves shifts in both image appearance and segmentation semantics with intricate correlation, causing concurrent appearance and semantic forgetting. To solve this issue, we propose a Comprehensive Generative Replay (CGR) framework that restores appearance and semantic knowledge by synthesizing image-mask pairs to mimic past task data, which focuses on two aspects: modeling image-mask correspondence and promoting scalability for diverse tasks. Specifically, we introduce a novel Bayesian Joint Diffusion (BJD) model for high-quality synthesis of image-mask pairs with their correspondence explicitly preserved by conditional denoising. Furthermore, we develop a Task-Oriented Adapter (TOA) that recalibrates prompt embeddings to modulate the diffusion model, making the data synthesis compatible with different tasks. Experiments on incremental tasks (cardiac, fundus and prostate segmentation) show its clear advantage for alleviating concurrent appearance and semantic forgetting. Code is available at https://github.com/jingyzhang/CGR.
A-Eval: A Benchmark for Cross-Dataset Evaluation of Abdominal Multi-Organ Segmentation
Sep 07, 2023



Abstract:Although deep learning have revolutionized abdominal multi-organ segmentation, models often struggle with generalization due to training on small, specific datasets. With the recent emergence of large-scale datasets, some important questions arise: \textbf{Can models trained on these datasets generalize well on different ones? If yes/no, how to further improve their generalizability?} To address these questions, we introduce A-Eval, a benchmark for the cross-dataset Evaluation ('Eval') of Abdominal ('A') multi-organ segmentation. We employ training sets from four large-scale public datasets: FLARE22, AMOS, WORD, and TotalSegmentator, each providing extensive labels for abdominal multi-organ segmentation. For evaluation, we incorporate the validation sets from these datasets along with the training set from the BTCV dataset, forming a robust benchmark comprising five distinct datasets. We evaluate the generalizability of various models using the A-Eval benchmark, with a focus on diverse data usage scenarios: training on individual datasets independently, utilizing unlabeled data via pseudo-labeling, mixing different modalities, and joint training across all available datasets. Additionally, we explore the impact of model sizes on cross-dataset generalizability. Through these analyses, we underline the importance of effective data usage in enhancing models' generalization capabilities, offering valuable insights for assembling large-scale datasets and improving training strategies. The code and pre-trained models are available at \href{https://github.com/uni-medical/A-Eval}{https://github.com/uni-medical/A-Eval}.
STU-Net: Scalable and Transferable Medical Image Segmentation Models Empowered by Large-Scale Supervised Pre-training
Apr 13, 2023



Abstract:Large-scale models pre-trained on large-scale datasets have profoundly advanced the development of deep learning. However, the state-of-the-art models for medical image segmentation are still small-scale, with their parameters only in the tens of millions. Further scaling them up to higher orders of magnitude is rarely explored. An overarching goal of exploring large-scale models is to train them on large-scale medical segmentation datasets for better transfer capacities. In this work, we design a series of Scalable and Transferable U-Net (STU-Net) models, with parameter sizes ranging from 14 million to 1.4 billion. Notably, the 1.4B STU-Net is the largest medical image segmentation model to date. Our STU-Net is based on nnU-Net framework due to its popularity and impressive performance. We first refine the default convolutional blocks in nnU-Net to make them scalable. Then, we empirically evaluate different scaling combinations of network depth and width, discovering that it is optimal to scale model depth and width together. We train our scalable STU-Net models on a large-scale TotalSegmentator dataset and find that increasing model size brings a stronger performance gain. This observation reveals that a large model is promising in medical image segmentation. Furthermore, we evaluate the transferability of our model on 14 downstream datasets for direct inference and 3 datasets for further fine-tuning, covering various modalities and segmentation targets. We observe good performance of our pre-trained model in both direct inference and fine-tuning. The code and pre-trained models are available at https://github.com/Ziyan-Huang/STU-Net.
A Novel Hybrid Convolutional Neural Network for Accurate Organ Segmentation in 3D Head and Neck CT Images
Sep 26, 2021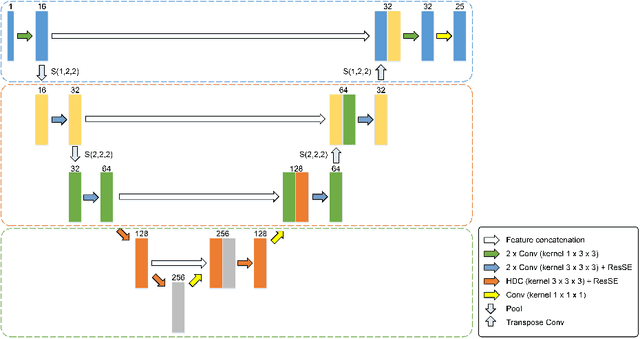

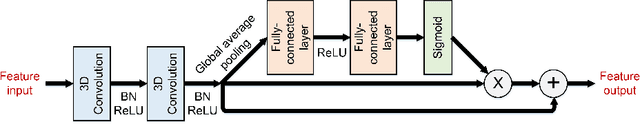
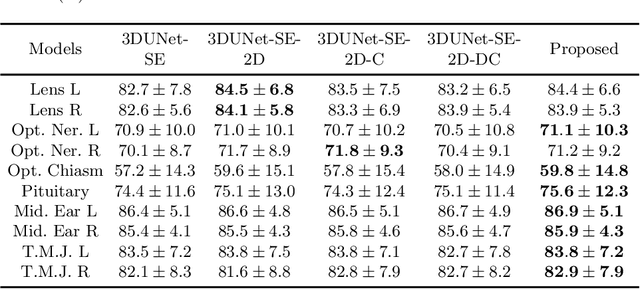
Abstract:Radiation therapy (RT) is widely employed in the clinic for the treatment of head and neck (HaN) cancers. An essential step of RT planning is the accurate segmentation of various organs-at-risks (OARs) in HaN CT images. Nevertheless, segmenting OARs manually is time-consuming, tedious, and error-prone considering that typical HaN CT images contain tens to hundreds of slices. Automated segmentation algorithms are urgently required. Recently, convolutional neural networks (CNNs) have been extensively investigated on this task. Particularly, 3D CNNs are frequently adopted to process 3D HaN CT images. There are two issues with na\"ive 3D CNNs. First, the depth resolution of 3D CT images is usually several times lower than the in-plane resolution. Direct employment of 3D CNNs without distinguishing this difference can lead to the extraction of distorted image features and influence the final segmentation performance. Second, a severe class imbalance problem exists, and large organs can be orders of times larger than small organs. It is difficult to simultaneously achieve accurate segmentation for all the organs. To address these issues, we propose a novel hybrid CNN that fuses 2D and 3D convolutions to combat the different spatial resolutions and extract effective edge and semantic features from 3D HaN CT images. To accommodate large and small organs, our final model, named OrganNet2.5D, consists of only two instead of the classic four downsampling operations, and hybrid dilated convolutions are introduced to maintain the respective field. Experiments on the MICCAI 2015 challenge dataset demonstrate that OrganNet2.5D achieves promising performance compared to state-of-the-art methods.
Group Shift Pointwise Convolution for Volumetric Medical Image Segmentation
Sep 26, 2021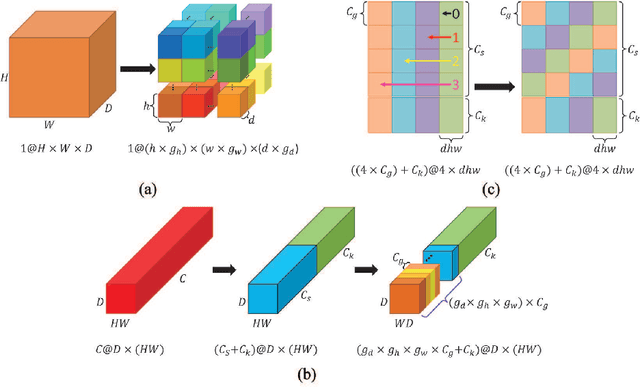
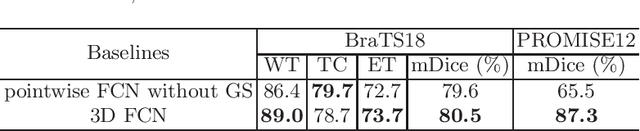
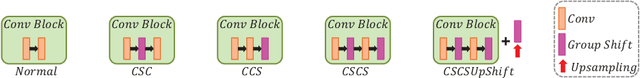
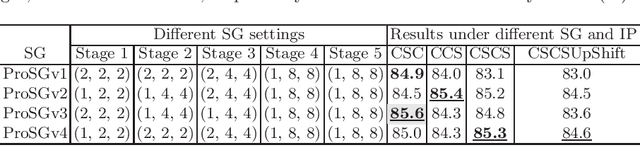
Abstract:Recent studies have witnessed the effectiveness of 3D convolutions on segmenting volumetric medical images. Compared with the 2D counterparts, 3D convolutions can capture the spatial context in three dimensions. Nevertheless, models employing 3D convolutions introduce more trainable parameters and are more computationally complex, which may lead easily to model overfitting especially for medical applications with limited available training data. This paper aims to improve the effectiveness and efficiency of 3D convolutions by introducing a novel Group Shift Pointwise Convolution (GSP-Conv). GSP-Conv simplifies 3D convolutions into pointwise ones with 1x1x1 kernels, which dramatically reduces the number of model parameters and FLOPs (e.g. 27x fewer than 3D convolutions with 3x3x3 kernels). Na\"ive pointwise convolutions with limited receptive fields cannot make full use of the spatial image context. To address this problem, we propose a parameter-free operation, Group Shift (GS), which shifts the feature maps along with different spatial directions in an elegant way. With GS, pointwise convolutions can access features from different spatial locations, and the limited receptive fields of pointwise convolutions can be compensated. We evaluate the proposed methods on two datasets, PROMISE12 and BraTS18. Results show that our method, with substantially decreased model complexity, achieves comparable or even better performance than models employing 3D convolutions.
SS-CADA: A Semi-Supervised Cross-Anatomy Domain Adaptation for Coronary Artery Segmentation
May 06, 2021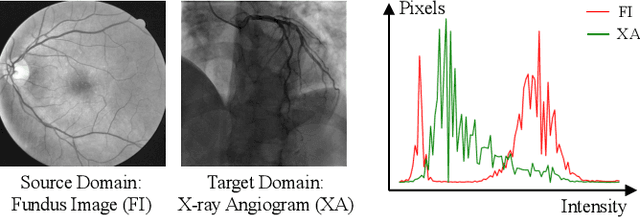
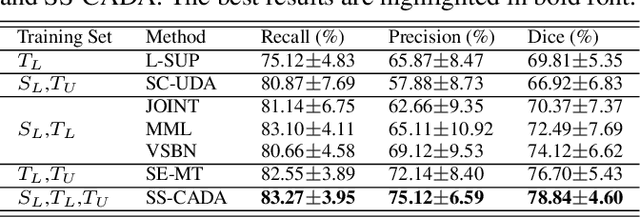

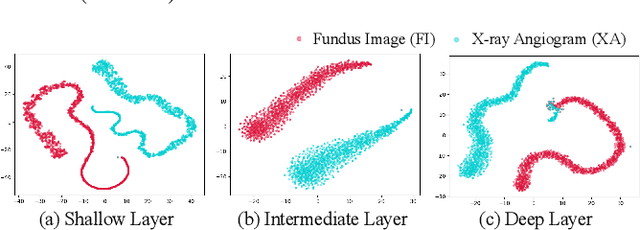
Abstract:The segmentation of coronary arteries by convolutional neural network is promising yet requires a large amount of labor-intensive manual annotations. Transferring knowledge from retinal vessels in widely-available public labeled fundus images (FIs) has a potential to reduce the annotation requirement for coronary artery segmentation in X-ray angiograms (XAs) due to their common tubular structures. However, it is challenged by the cross-anatomy domain shift due to the intrinsically different vesselness characteristics in different anatomical regions under even different imaging protocols. To solve this problem, we propose a Semi-Supervised Cross-Anatomy Domain Adaptation (SS-CADA) which requires only limited annotations for coronary arteries in XAs. With the supervision from a small number of labeled XAs and publicly available labeled FIs, we propose a vesselness-specific batch normalization (VSBN) to individually normalize feature maps for them considering their different cross-anatomic vesselness characteristics. In addition, to further facilitate the annotation efficiency, we employ a self-ensembling mean-teacher (SEMT) to exploit abundant unlabeled XAs by imposing a prediction consistency constraint. Extensive experiments show that our SS-CADA is able to solve the challenging cross-anatomy domain shift, achieving accurate segmentation for coronary arteries given only a small number of labeled XAs.
Weakly Supervised Vessel Segmentation in X-ray Angiograms by Self-Paced Learning from Noisy Labels with Suggestive Annotation
May 27, 2020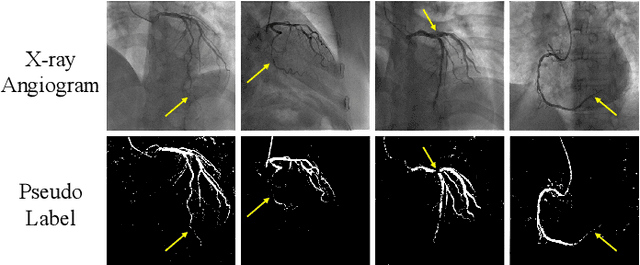
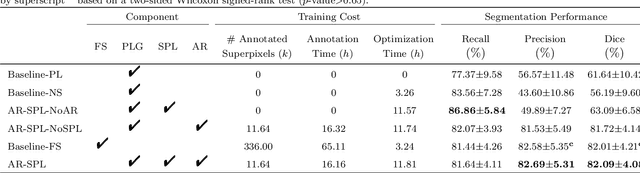
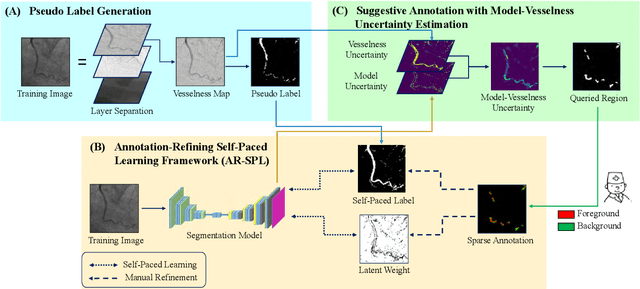
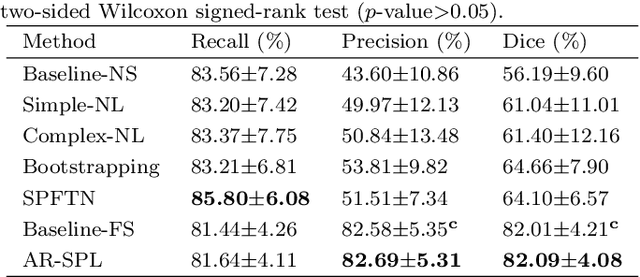
Abstract:The segmentation of coronary arteries in X-ray angiograms by convolutional neural networks (CNNs) is promising yet limited by the requirement of precisely annotating all pixels in a large number of training images, which is extremely labor-intensive especially for complex coronary trees. To alleviate the burden on the annotator, we propose a novel weakly supervised training framework that learns from noisy pseudo labels generated from automatic vessel enhancement, rather than accurate labels obtained by fully manual annotation. A typical self-paced learning scheme is used to make the training process robust against label noise while challenged by the systematic biases in pseudo labels, thus leading to the decreased performance of CNNs at test time. To solve this problem, we propose an annotation-refining self-paced learning framework (AR-SPL) to correct the potential errors using suggestive annotation. An elaborate model-vesselness uncertainty estimation is also proposed to enable the minimal annotation cost for suggestive annotation, based on not only the CNNs in training but also the geometric features of coronary arteries derived directly from raw data. Experiments show that our proposed framework achieves 1) comparable accuracy to fully supervised learning, which also significantly outperforms other weakly supervised learning frameworks; 2) largely reduced annotation cost, i.e., 75.18% of annotation time is saved, and only 3.46% of image regions are required to be annotated; and 3) an efficient intervention process, leading to superior performance with even fewer manual interactions.
A novel active learning framework for classification: using weighted rank aggregation to achieve multiple query criteria
Sep 27, 2018



Abstract:Multiple query criteria active learning (MQCAL) methods have a higher potential performance than conventional active learning methods in which only one criterion is deployed for sample selection. A central issue related to MQCAL methods concerns the development of an integration criteria strategy (ICS) that makes full use of all criteria. The conventional ICS adopted in relevant research all facilitate the desired effects, but several limitations still must be addressed. For instance, some of the strategies are not sufficiently scalable during the design process, and the number and type of criteria involved are dictated. Thus, it is challenging for the user to integrate other criteria into the original process unless modifications are made to the algorithm. Other strategies are too dependent on empirical parameters, which can only be acquired by experience or cross-validation and thus lack generality; additionally, these strategies are counter to the intention of active learning, as samples need to be labeled in the validation set before the active learning process can begin. To address these limitations, we propose a novel MQCAL method for classification tasks that employs a third strategy via weighted rank aggregation. The proposed method serves as a heuristic means to select high-value samples of high scalability and generality and is implemented through a three-step process: (1) the transformation of the sample selection to sample ranking and scoring, (2) the computation of the self-adaptive weights of each criterion, and (3) the weighted aggregation of each sample rank list. Ultimately, the sample at the top of the aggregated ranking list is the most comprehensively valuable and must be labeled. Several experiments generating 257 wins, 194 ties and 49 losses against other state-of-the-art MQCALs are conducted to verify that the proposed method can achieve superior results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge