Hui Sun
ChatAD: Reasoning-Enhanced Time-Series Anomaly Detection with Multi-Turn Instruction Evolution
Jan 20, 2026Abstract:LLM-driven Anomaly Detection (AD) helps enhance the understanding and explanatory abilities of anomalous behaviors in Time Series (TS). Existing methods face challenges of inadequate reasoning ability, deficient multi-turn dialogue capability, and narrow generalization. To this end, we 1) propose a multi-agent-based TS Evolution algorithm named TSEvol. On top of it, we 2) introduce the AD reasoning and multi-turn dialogue Dataset TSEData-20K and contribute the Chatbot family for AD, including ChatAD-Llama3-8B, Qwen2.5-7B, and Mistral-7B. Furthermore, 3) we propose the TS Kahneman-Tversky Optimization (TKTO) to enhance ChatAD's cross-task generalization capability. Lastly, 4) we propose a LLM-driven Learning-based AD Benchmark LLADBench to evaluate the performance of ChatAD and nine baselines across seven datasets and tasks. Our three ChatAD models achieve substantial gains, up to 34.50% in accuracy, 34.71% in F1, and a 37.42% reduction in false positives. Besides, via KTKO, our optimized ChatAD achieves competitive performance in reasoning and cross-task generalization on classification, forecasting, and imputation.
The Ensemble Schr{ö}dinger Bridge filter for Nonlinear Data Assimilation
Dec 22, 2025Abstract:This work puts forward a novel nonlinear optimal filter namely the Ensemble Schr{ö}dinger Bridge nonlinear filter. The proposed filter finds marriage of the standard prediction procedure and the diffusion generative modeling for the analysis procedure to realize one filtering step. The designed approach finds no structural model error, and it is derivative free, training free and highly parallizable. Experimental results show that the designed algorithm performs well given highly nonlinear dynamics in (mildly) high dimension up to 40 or above under a chaotic environment. It also shows better performance than classical methods such as the ensemble Kalman filter and the Particle filter in numerous tests given different level of nonlinearity. Future work will focus on extending the proposed approach to practical meteorological applications and establishing a rigorous convergence analysis.
Spatial-Frequency Enhanced Mamba for Multi-Modal Image Fusion
Nov 10, 2025Abstract:Multi-Modal Image Fusion (MMIF) aims to integrate complementary image information from different modalities to produce informative images. Previous deep learning-based MMIF methods generally adopt Convolutional Neural Networks (CNNs) or Transformers for feature extraction. However, these methods deliver unsatisfactory performances due to the limited receptive field of CNNs and the high computational cost of Transformers. Recently, Mamba has demonstrated a powerful potential for modeling long-range dependencies with linear complexity, providing a promising solution to MMIF. Unfortunately, Mamba lacks full spatial and frequency perceptions, which are very important for MMIF. Moreover, employing Image Reconstruction (IR) as an auxiliary task has been proven beneficial for MMIF. However, a primary challenge is how to leverage IR efficiently and effectively. To address the above issues, we propose a novel framework named Spatial-Frequency Enhanced Mamba Fusion (SFMFusion) for MMIF. More specifically, we first propose a three-branch structure to couple MMIF and IR, which can retain complete contents from source images. Then, we propose the Spatial-Frequency Enhanced Mamba Block (SFMB), which can enhance Mamba in both spatial and frequency domains for comprehensive feature extraction. Finally, we propose the Dynamic Fusion Mamba Block (DFMB), which can be deployed across different branches for dynamic feature fusion. Extensive experiments show that our method achieves better results than most state-of-the-art methods on six MMIF datasets. The source code is available at https://github.com/SunHui1216/SFMFusion.
OneTrans: Unified Feature Interaction and Sequence Modeling with One Transformer in Industrial Recommender
Oct 30, 2025Abstract:In recommendation systems, scaling up feature-interaction modules (e.g., Wukong, RankMixer) or user-behavior sequence modules (e.g., LONGER) has achieved notable success. However, these efforts typically proceed on separate tracks, which not only hinders bidirectional information exchange but also prevents unified optimization and scaling. In this paper, we propose OneTrans, a unified Transformer backbone that simultaneously performs user-behavior sequence modeling and feature interaction. OneTrans employs a unified tokenizer to convert both sequential and non-sequential attributes into a single token sequence. The stacked OneTrans blocks share parameters across similar sequential tokens while assigning token-specific parameters to non-sequential tokens. Through causal attention and cross-request KV caching, OneTrans enables precomputation and caching of intermediate representations, significantly reducing computational costs during both training and inference. Experimental results on industrial-scale datasets demonstrate that OneTrans scales efficiently with increasing parameters, consistently outperforms strong baselines, and yields a 5.68% lift in per-user GMV in online A/B tests.
NetSight: Graph Attention Based Traffic Forecasting in Computer Networks
May 11, 2025Abstract:The traffic in today's networks is increasingly influenced by the interactions among network nodes as well as by the temporal fluctuations in the demands of the nodes. Traditional statistical prediction methods are becoming obsolete due to their inability to address the non-linear and dynamic spatio-temporal dependencies present in today's network traffic. The most promising direction of research today is graph neural networks (GNNs) based prediction approaches that are naturally suited to handle graph-structured data. Unfortunately, the state-of-the-art GNN approaches separate the modeling of spatial and temporal information, resulting in the loss of important information about joint dependencies. These GNN based approaches further do not model information at both local and global scales simultaneously, leaving significant room for improvement. To address these challenges, we propose NetSight. NetSight learns joint spatio-temporal dependencies simultaneously at both global and local scales from the time-series of measurements of any given network metric collected at various nodes in a network. Using the learned information, NetSight can then accurately predict the future values of the given network metric at those nodes in the network. We propose several new concepts and techniques in the design of NetSight, such as spatio-temporal adjacency matrix and node normalization. Through extensive evaluations and comparison with prior approaches using data from two large real-world networks, we show that NetSight significantly outperforms all prior state-of-the-art approaches. We will release the source code and data used in the evaluation of NetSight on the acceptance of this paper.
Post-Incorporating Code Structural Knowledge into LLMs via In-Context Learning for Code Translation
Mar 28, 2025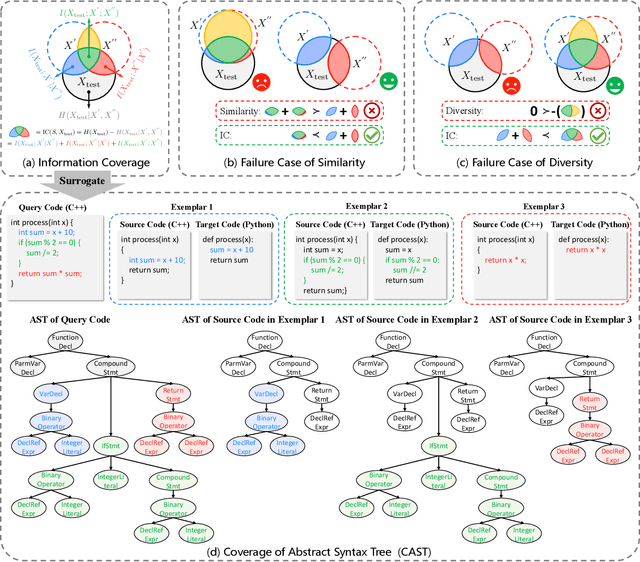
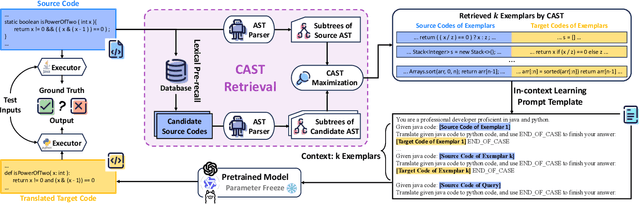
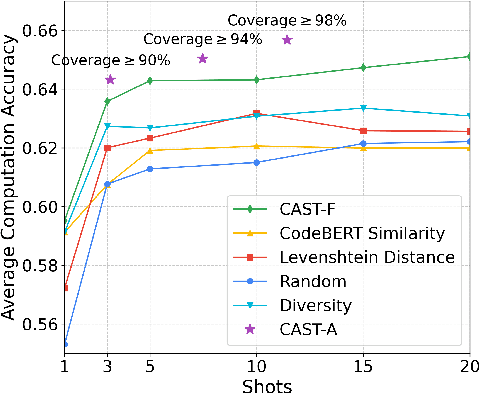
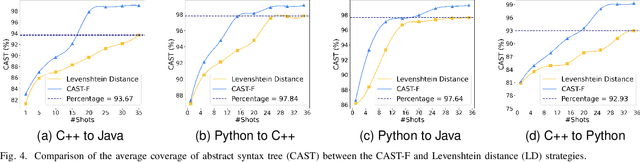
Abstract:Code translation migrates codebases across programming languages. Recently, large language models (LLMs) have achieved significant advancements in software mining. However, handling the syntactic structure of source code remains a challenge. Classic syntax-aware methods depend on intricate model architectures and loss functions, rendering their integration into LLM training resource-intensive. This paper employs in-context learning (ICL), which directly integrates task exemplars into the input context, to post-incorporate code structural knowledge into pre-trained LLMs. We revisit exemplar selection in ICL from an information-theoretic perspective, proposing that list-wise selection based on information coverage is more precise and general objective than traditional methods based on combining similarity and diversity. To address the challenges of quantifying information coverage, we introduce a surrogate measure, Coverage of Abstract Syntax Tree (CAST). Furthermore, we formulate the NP-hard CAST maximization for exemplar selection and prove that it is a standard submodular maximization problem. Therefore, we propose a greedy algorithm for CAST submodular maximization, which theoretically guarantees a (1-1/e)-approximate solution in polynomial time complexity. Our method is the first training-free and model-agnostic approach to post-incorporate code structural knowledge into existing LLMs at test time. Experimental results show that our method significantly improves LLMs performance and reveals two meaningful insights: 1) Code structural knowledge can be effectively post-incorporated into pre-trained LLMs during inference, despite being overlooked during training; 2) Scaling up model size or training data does not lead to the emergence of code structural knowledge, underscoring the necessity of explicitly considering code syntactic structure.
MDP3: A Training-free Approach for List-wise Frame Selection in Video-LLMs
Jan 06, 2025Abstract:Video large language models (Video-LLMs) have made significant progress in understanding videos. However, processing multiple frames leads to lengthy visual token sequences, presenting challenges such as the limited context length cannot accommodate the entire video, and the inclusion of irrelevant frames hinders visual perception. Hence, effective frame selection is crucial. This paper emphasizes that frame selection should follow three key principles: query relevance, list-wise diversity, and sequentiality. Existing methods, such as uniform frame sampling and query-frame matching, do not capture all of these principles. Thus, we propose Markov decision determinantal point process with dynamic programming (MDP3) for frame selection, a training-free and model-agnostic method that can be seamlessly integrated into existing Video-LLMs. Our method first estimates frame similarities conditioned on the query using a conditional Gaussian kernel within the reproducing kernel Hilbert space~(RKHS). We then apply the determinantal point process~(DPP) to the similarity matrix to capture both query relevance and list-wise diversity. To incorporate sequentiality, we segment the video and apply DPP within each segment, conditioned on the preceding segment selection, modeled as a Markov decision process~(MDP) for allocating selection sizes across segments. Theoretically, MDP3 provides a \((1 - 1/e)\)-approximate solution to the NP-hard list-wise frame selection problem with pseudo-polynomial time complexity, demonstrating its efficiency. Empirically, MDP3 significantly outperforms existing methods, verifying its effectiveness and robustness.
A Joint Learning Model with Variational Interaction for Multilingual Program Translation
Aug 25, 2024

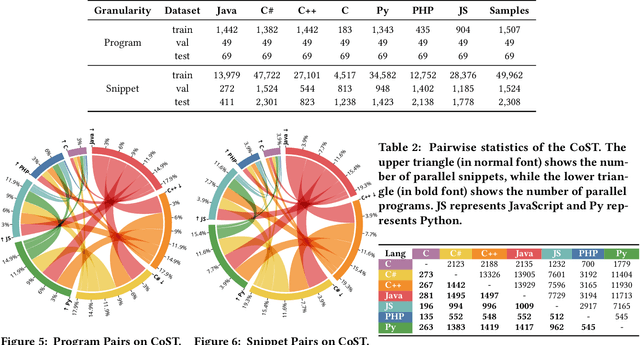

Abstract:Programs implemented in various programming languages form the foundation of software applications. To alleviate the burden of program migration and facilitate the development of software systems, automated program translation across languages has garnered significant attention. Previous approaches primarily focus on pairwise translation paradigms, learning translation between pairs of languages using bilingual parallel data. However, parallel data is difficult to collect for some language pairs, and the distribution of program semantics across languages can shift, posing challenges for pairwise program translation. In this paper, we argue that jointly learning a unified model to translate code across multiple programming languages is superior to separately learning from bilingual parallel data. We propose Variational Interaction for Multilingual Program Translation~(VIM-PT), a disentanglement-based generative approach that jointly trains a unified model for multilingual program translation across multiple languages. VIM-PT disentangles code into language-shared and language-specific features, using variational inference and interaction information with a novel lower bound, then achieves program translation through conditional generation. VIM-PT demonstrates four advantages: 1) captures language-shared information more accurately from various implementations and improves the quality of multilingual program translation, 2) mines and leverages the capability of non-parallel data, 3) addresses the distribution shift of program semantics across languages, 4) and serves as a unified model, reducing deployment complexity.
Large Language Models for Link Stealing Attacks Against Graph Neural Networks
Jun 22, 2024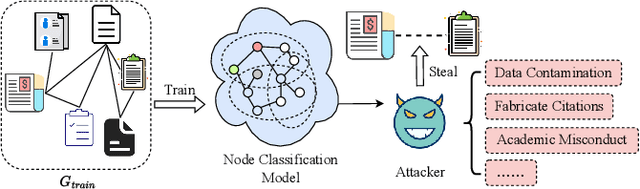



Abstract:Graph data contains rich node features and unique edge information, which have been applied across various domains, such as citation networks or recommendation systems. Graph Neural Networks (GNNs) are specialized for handling such data and have shown impressive performance in many applications. However, GNNs may contain of sensitive information and susceptible to privacy attacks. For example, link stealing is a type of attack in which attackers infer whether two nodes are linked or not. Previous link stealing attacks primarily relied on posterior probabilities from the target GNN model, neglecting the significance of node features. Additionally, variations in node classes across different datasets lead to different dimensions of posterior probabilities. The handling of these varying data dimensions posed a challenge in using a single model to effectively conduct link stealing attacks on different datasets. To address these challenges, we introduce Large Language Models (LLMs) to perform link stealing attacks on GNNs. LLMs can effectively integrate textual features and exhibit strong generalizability, enabling attacks to handle diverse data dimensions across various datasets. We design two distinct LLM prompts to effectively combine textual features and posterior probabilities of graph nodes. Through these designed prompts, we fine-tune the LLM to adapt to the link stealing attack task. Furthermore, we fine-tune the LLM using multiple datasets and enable the LLM to learn features from different datasets simultaneously. Experimental results show that our approach significantly enhances the performance of existing link stealing attack tasks in both white-box and black-box scenarios. Our method can execute link stealing attacks across different datasets using only a single model, making link stealing attacks more applicable to real-world scenarios.
FViT: A Focal Vision Transformer with Gabor Filter
Feb 27, 2024Abstract:Vision transformers have achieved encouraging progress in various computer vision tasks. A common belief is that this is attributed to the competence of self-attention in modeling the global dependencies among feature tokens. Unfortunately, self-attention still faces some challenges in dense prediction tasks, such as the high computational complexity and absence of desirable inductive bias. To address these issues, we revisit the potential benefits of integrating vision transformer with Gabor filter, and propose a Learnable Gabor Filter (LGF) by using convolution. As an alternative to self-attention, we employ LGF to simulate the response of simple cells in the biological visual system to input images, prompting models to focus on discriminative feature representations of targets from various scales and orientations. Additionally, we design a Bionic Focal Vision (BFV) block based on the LGF. This block draws inspiration from neuroscience and introduces a Multi-Path Feed Forward Network (MPFFN) to emulate the working way of biological visual cortex processing information in parallel. Furthermore, we develop a unified and efficient pyramid backbone network family called Focal Vision Transformers (FViTs) by stacking BFV blocks. Experimental results show that FViTs exhibit highly competitive performance in various vision tasks. Especially in terms of computational efficiency and scalability, FViTs show significant advantages compared with other counterparts. Code is available at https://github.com/nkusyl/FViT
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge