Lianqing Liu
SeqWalker: Sequential-Horizon Vision-and-Language Navigation with Hierarchical Planning
Jan 08, 2026Abstract:Sequential-Horizon Vision-and-Language Navigation (SH-VLN) presents a challenging scenario where agents should sequentially execute multi-task navigation guided by complex, long-horizon language instructions. Current vision-and-language navigation models exhibit significant performance degradation with such multi-task instructions, as information overload impairs the agent's ability to attend to observationally relevant details. To address this problem, we propose SeqWalker, a navigation model built on a hierarchical planning framework. Our SeqWalker features: i) A High-Level Planner that dynamically selects global instructions into contextually relevant sub-instructions based on the agent's current visual observations, thus reducing cognitive load; ii) A Low-Level Planner incorporating an Exploration-Verification strategy that leverages the inherent logical structure of instructions for trajectory error correction. To evaluate SH-VLN performance, we also extend the IVLN dataset and establish a new benchmark. Extensive experiments are performed to demonstrate the superiority of the proposed SeqWalker.
CRISP: Contrastive Residual Injection and Semantic Prompting for Continual Video Instance Segmentation
Aug 14, 2025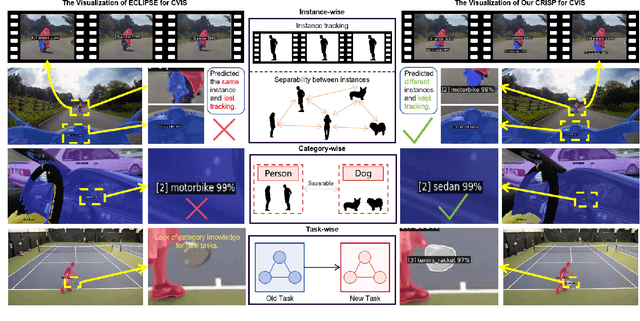
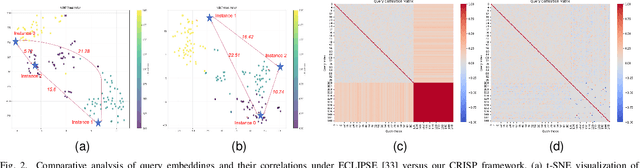
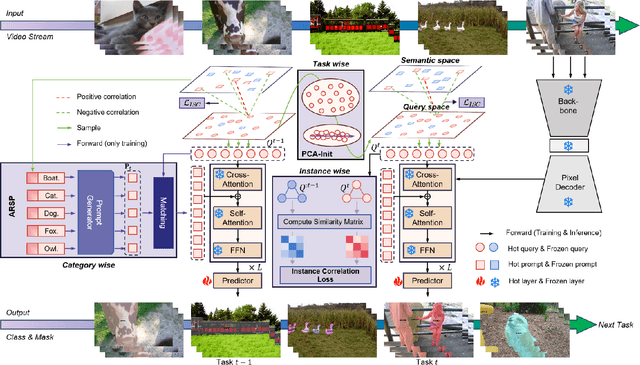
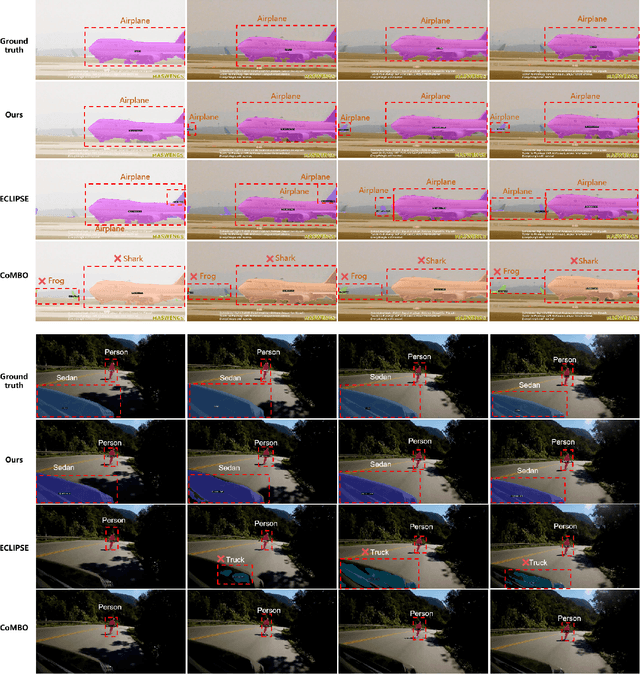
Abstract:Continual video instance segmentation demands both the plasticity to absorb new object categories and the stability to retain previously learned ones, all while preserving temporal consistency across frames. In this work, we introduce Contrastive Residual Injection and Semantic Prompting (CRISP), an earlier attempt tailored to address the instance-wise, category-wise, and task-wise confusion in continual video instance segmentation. For instance-wise learning, we model instance tracking and construct instance correlation loss, which emphasizes the correlation with the prior query space while strengthening the specificity of the current task query. For category-wise learning, we build an adaptive residual semantic prompt (ARSP) learning framework, which constructs a learnable semantic residual prompt pool generated by category text and uses an adjustive query-prompt matching mechanism to build a mapping relationship between the query of the current task and the semantic residual prompt. Meanwhile, a semantic consistency loss based on the contrastive learning is introduced to maintain semantic coherence between object queries and residual prompts during incremental training. For task-wise learning, to ensure the correlation at the inter-task level within the query space, we introduce a concise yet powerful initialization strategy for incremental prompts. Extensive experiments on YouTube-VIS-2019 and YouTube-VIS-2021 datasets demonstrate that CRISP significantly outperforms existing continual segmentation methods in the long-term continual video instance segmentation task, avoiding catastrophic forgetting and effectively improving segmentation and classification performance. The code is available at https://github.com/01upup10/CRISP.
K2MUSE: A human lower limb multimodal dataset under diverse conditions for facilitating rehabilitation robotics
Apr 20, 2025Abstract:The natural interaction and control performance of lower limb rehabilitation robots are closely linked to biomechanical information from various human locomotion activities. Multidimensional human motion data significantly deepen the understanding of the complex mechanisms governing neuromuscular alterations, thereby facilitating the development and application of rehabilitation robots in multifaceted real-world environments. However, currently available lower limb datasets are inadequate for supplying the essential multimodal data and large-scale gait samples necessary for effective data-driven approaches, and they neglect the significant effects of acquisition interference in real applications.To fill this gap, we present the K2MUSE dataset, which includes a comprehensive collection of multimodal data, comprising kinematic, kinetic, amplitude-mode ultrasound (AUS), and surface electromyography (sEMG) measurements. The proposed dataset includes lower limb multimodal data from 30 able-bodied participants walking under different inclines (0$^\circ$, $\pm$5$^\circ$, and $\pm$10$^\circ$), various speeds (0.5 m/s, 1.0 m/s, and 1.5 m/s), and different nonideal acquisition conditions (muscle fatigue, electrode shifts, and inter-day differences). The kinematic and ground reaction force data were collected via a Vicon motion capture system and an instrumented treadmill with embedded force plates, whereas the sEMG and AUS data were synchronously recorded for thirteen muscles on the bilateral lower limbs. This dataset offers a new resource for designing control frameworks for rehabilitation robots and conducting biomechanical analyses of lower limb locomotion. The dataset is available at https://k2muse.github.io/.
CAD-Mesher: A Convenient, Accurate, Dense Mesh-based Mapping Module in SLAM for Dynamic Environments
Aug 12, 2024Abstract:Most LiDAR odometry and SLAM systems construct maps in point clouds, which are discrete and sparse when zoomed in, making them not directly suitable for navigation. Mesh maps represent a dense and continuous map format with low memory consumption, which can approximate complex structures with simple elements, attracting significant attention of researchers in recent years. However, most implementations operate under a static environment assumption. In effect, moving objects cause ghosting, potentially degrading the quality of meshing. To address these issues, we propose a plug-and-play meshing module adapting to dynamic environments, which can easily integrate with various LiDAR odometry to generally improve the pose estimation accuracy of odometry. In our meshing module, a novel two-stage coarse-to-fine dynamic removal method is designed to effectively filter dynamic objects, generating consistent, accurate, and dense mesh maps. To our best know, this is the first mesh construction method with explicit dynamic removal. Additionally, conducive to Gaussian process in mesh construction, sliding window-based keyframe aggregation and adaptive downsampling strategies are used to ensure the uniformity of point cloud. We evaluate the localization and mapping accuracy on five publicly available datasets. Both qualitative and quantitative results demonstrate the superiority of our method compared with the state-of-the-art algorithms. The code and introduction video are publicly available at https://yaepiii.github.io/CAD-Mesher/.
Efficient Model Learning and Adaptive Tracking Control of Magnetic Micro-Robots for Non-Contact Manipulation
Mar 21, 2024



Abstract:Magnetic microrobots can be navigated by an external magnetic field to autonomously move within living organisms with complex and unstructured environments. Potential applications include drug delivery, diagnostics, and therapeutic interventions. Existing techniques commonly impart magnetic properties to the target object,or drive the robot to contact and then manipulate the object, both probably inducing physical damage. This paper considers a non-contact formulation, where the robot spins to generate a repulsive field to push the object without physical contact. Under such a formulation, the main challenge is that the motion model between the input of the magnetic field and the output velocity of the target object is commonly unknown and difficult to analyze. To deal with it, this paper proposes a data-driven-based solution. A neural network is constructed to efficiently estimate the motion model. Then, an approximate model-based optimal control scheme is developed to push the object to track a time-varying trajectory, maintaining the non-contact with distance constraints. Furthermore, a straightforward planner is introduced to assess the adaptability of non-contact manipulation in a cluttered unstructured environment. Experimental results are presented to show the tracking and navigation performance of the proposed scheme.
Deep Learning Enables Large Depth-of-Field Images for Sub-Diffraction-Limit Scanning Superlens Microscopy
Oct 27, 2023



Abstract:Scanning electron microscopy (SEM) is indispensable in diverse applications ranging from microelectronics to food processing because it provides large depth-of-field images with a resolution beyond the optical diffraction limit. However, the technology requires coating conductive films on insulator samples and a vacuum environment. We use deep learning to obtain the mapping relationship between optical super-resolution (OSR) images and SEM domain images, which enables the transformation of OSR images into SEM-like large depth-of-field images. Our custom-built scanning superlens microscopy (SSUM) system, which requires neither coating samples by conductive films nor a vacuum environment, is used to acquire the OSR images with features down to ~80 nm. The peak signal-to-noise ratio (PSNR) and structural similarity index measure values indicate that the deep learning method performs excellently in image-to-image translation, with a PSNR improvement of about 0.74 dB over the optical super-resolution images. The proposed method provides a high level of detail in the reconstructed results, indicating that it has broad applicability to chip-level defect detection, biological sample analysis, forensics, and various other fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge