Chenglong Ma
Probing Scientific General Intelligence of LLMs with Scientist-Aligned Workflows
Dec 18, 2025Abstract:Despite advances in scientific AI, a coherent framework for Scientific General Intelligence (SGI)-the ability to autonomously conceive, investigate, and reason across scientific domains-remains lacking. We present an operational SGI definition grounded in the Practical Inquiry Model (PIM: Deliberation, Conception, Action, Perception) and operationalize it via four scientist-aligned tasks: deep research, idea generation, dry/wet experiments, and experimental reasoning. SGI-Bench comprises over 1,000 expert-curated, cross-disciplinary samples inspired by Science's 125 Big Questions, enabling systematic evaluation of state-of-the-art LLMs. Results reveal gaps: low exact match (10--20%) in deep research despite step-level alignment; ideas lacking feasibility and detail; high code executability but low execution result accuracy in dry experiments; low sequence fidelity in wet protocols; and persistent multimodal comparative-reasoning challenges. We further introduce Test-Time Reinforcement Learning (TTRL), which optimizes retrieval-augmented novelty rewards at inference, enhancing hypothesis novelty without reference answer. Together, our PIM-grounded definition, workflow-centric benchmark, and empirical insights establish a foundation for AI systems that genuinely participate in scientific discovery.
MedQ-Bench: Evaluating and Exploring Medical Image Quality Assessment Abilities in MLLMs
Oct 02, 2025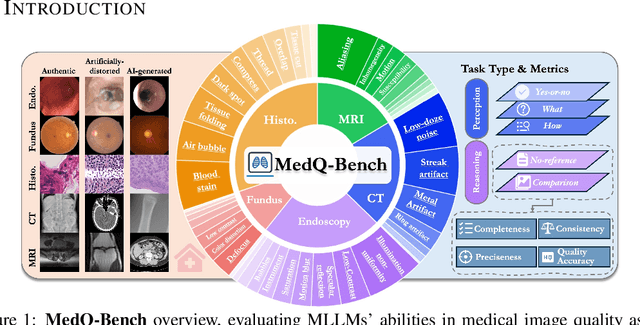
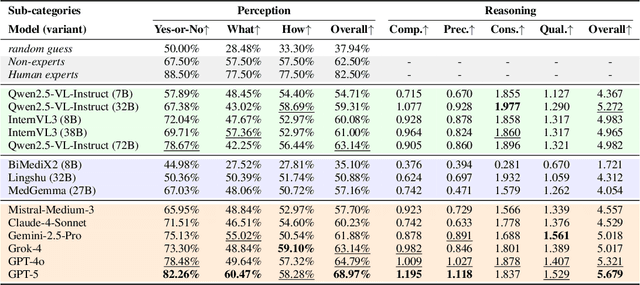
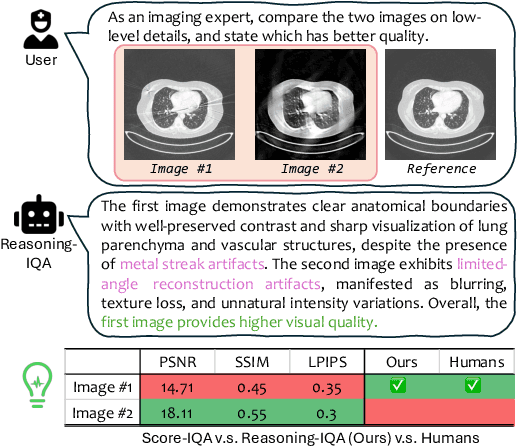
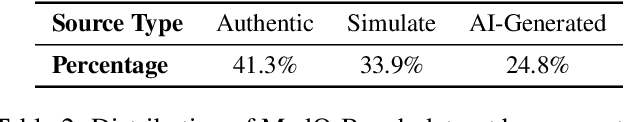
Abstract:Medical Image Quality Assessment (IQA) serves as the first-mile safety gate for clinical AI, yet existing approaches remain constrained by scalar, score-based metrics and fail to reflect the descriptive, human-like reasoning process central to expert evaluation. To address this gap, we introduce MedQ-Bench, a comprehensive benchmark that establishes a perception-reasoning paradigm for language-based evaluation of medical image quality with Multi-modal Large Language Models (MLLMs). MedQ-Bench defines two complementary tasks: (1) MedQ-Perception, which probes low-level perceptual capability via human-curated questions on fundamental visual attributes; and (2) MedQ-Reasoning, encompassing both no-reference and comparison reasoning tasks, aligning model evaluation with human-like reasoning on image quality. The benchmark spans five imaging modalities and over forty quality attributes, totaling 2,600 perceptual queries and 708 reasoning assessments, covering diverse image sources including authentic clinical acquisitions, images with simulated degradations via physics-based reconstructions, and AI-generated images. To evaluate reasoning ability, we propose a multi-dimensional judging protocol that assesses model outputs along four complementary axes. We further conduct rigorous human-AI alignment validation by comparing LLM-based judgement with radiologists. Our evaluation of 14 state-of-the-art MLLMs demonstrates that models exhibit preliminary but unstable perceptual and reasoning skills, with insufficient accuracy for reliable clinical use. These findings highlight the need for targeted optimization of MLLMs in medical IQA. We hope that MedQ-Bench will catalyze further exploration and unlock the untapped potential of MLLMs for medical image quality evaluation.
A Survey of Scientific Large Language Models: From Data Foundations to Agent Frontiers
Aug 28, 2025



Abstract:Scientific Large Language Models (Sci-LLMs) are transforming how knowledge is represented, integrated, and applied in scientific research, yet their progress is shaped by the complex nature of scientific data. This survey presents a comprehensive, data-centric synthesis that reframes the development of Sci-LLMs as a co-evolution between models and their underlying data substrate. We formulate a unified taxonomy of scientific data and a hierarchical model of scientific knowledge, emphasizing the multimodal, cross-scale, and domain-specific challenges that differentiate scientific corpora from general natural language processing datasets. We systematically review recent Sci-LLMs, from general-purpose foundations to specialized models across diverse scientific disciplines, alongside an extensive analysis of over 270 pre-/post-training datasets, showing why Sci-LLMs pose distinct demands -- heterogeneous, multi-scale, uncertainty-laden corpora that require representations preserving domain invariance and enabling cross-modal reasoning. On evaluation, we examine over 190 benchmark datasets and trace a shift from static exams toward process- and discovery-oriented assessments with advanced evaluation protocols. These data-centric analyses highlight persistent issues in scientific data development and discuss emerging solutions involving semi-automated annotation pipelines and expert validation. Finally, we outline a paradigm shift toward closed-loop systems where autonomous agents based on Sci-LLMs actively experiment, validate, and contribute to a living, evolving knowledge base. Collectively, this work provides a roadmap for building trustworthy, continually evolving artificial intelligence (AI) systems that function as a true partner in accelerating scientific discovery.
S2-UniSeg: Fast Universal Agglomerative Pooling for Scalable Segment Anything without Supervision
Aug 09, 2025Abstract:Recent self-supervised image segmentation models have achieved promising performance on semantic segmentation and class-agnostic instance segmentation. However, their pretraining schedule is multi-stage, requiring a time-consuming pseudo-masks generation process between each training epoch. This time-consuming offline process not only makes it difficult to scale with training dataset size, but also leads to sub-optimal solutions due to its discontinuous optimization routine. To solve these, we first present a novel pseudo-mask algorithm, Fast Universal Agglomerative Pooling (UniAP). Each layer of UniAP can identify groups of similar nodes in parallel, allowing to generate both semantic-level and instance-level and multi-granular pseudo-masks within ens of milliseconds for one image. Based on the fast UniAP, we propose the Scalable Self-Supervised Universal Segmentation (S2-UniSeg), which employs a student and a momentum teacher for continuous pretraining. A novel segmentation-oriented pretext task, Query-wise Self-Distillation (QuerySD), is proposed to pretrain S2-UniSeg to learn the local-to-global correspondences. Under the same setting, S2-UniSeg outperforms the SOTA UnSAM model, achieving notable improvements of AP+6.9 on COCO, AR+11.1 on UVO, PixelAcc+4.5 on COCOStuff-27, RQ+8.0 on Cityscapes. After scaling up to a larger 2M-image subset of SA-1B, S2-UniSeg further achieves performance gains on all four benchmarks. Our code and pretrained models are available at https://github.com/bio-mlhui/S2-UniSeg
PUB: An LLM-Enhanced Personality-Driven User Behaviour Simulator for Recommender System Evaluation
Jun 05, 2025Abstract:Traditional offline evaluation methods for recommender systems struggle to capture the complexity of modern platforms due to sparse behavioural signals, noisy data, and limited modelling of user personality traits. While simulation frameworks can generate synthetic data to address these gaps, existing methods fail to replicate behavioural diversity, limiting their effectiveness. To overcome these challenges, we propose the Personality-driven User Behaviour Simulator (PUB), an LLM-based simulation framework that integrates the Big Five personality traits to model personalised user behaviour. PUB dynamically infers user personality from behavioural logs (e.g., ratings, reviews) and item metadata, then generates synthetic interactions that preserve statistical fidelity to real-world data. Experiments on the Amazon review datasets show that logs generated by PUB closely align with real user behaviour and reveal meaningful associations between personality traits and recommendation outcomes. These results highlight the potential of the personality-driven simulator to advance recommender system evaluation, offering scalable, controllable, high-fidelity alternatives to resource-intensive real-world experiments.
MedITok: A Unified Tokenizer for Medical Image Synthesis and Interpretation
May 25, 2025Abstract:Advanced autoregressive models have reshaped multimodal AI. However, their transformative potential in medical imaging remains largely untapped due to the absence of a unified visual tokenizer -- one capable of capturing fine-grained visual structures for faithful image reconstruction and realistic image synthesis, as well as rich semantics for accurate diagnosis and image interpretation. To this end, we present MedITok, the first unified tokenizer tailored for medical images, encoding both low-level structural details and high-level clinical semantics within a unified latent space. To balance these competing objectives, we introduce a novel two-stage training framework: a visual representation alignment stage that cold-starts the tokenizer reconstruction learning with a visual semantic constraint, followed by a textual semantic representation alignment stage that infuses detailed clinical semantics into the latent space. Trained on the meticulously collected large-scale dataset with over 30 million medical images and 2 million image-caption pairs, MedITok achieves state-of-the-art performance on more than 30 datasets across 9 imaging modalities and 4 different tasks. By providing a unified token space for autoregressive modeling, MedITok supports a wide range of tasks in clinical diagnostics and generative healthcare applications. Model and code will be made publicly available at: https://github.com/Masaaki-75/meditok.
GMAI-VL-R1: Harnessing Reinforcement Learning for Multimodal Medical Reasoning
Apr 02, 2025Abstract:Recent advances in general medical AI have made significant strides, but existing models often lack the reasoning capabilities needed for complex medical decision-making. This paper presents GMAI-VL-R1, a multimodal medical reasoning model enhanced by reinforcement learning (RL) to improve its reasoning abilities. Through iterative training, GMAI-VL-R1 optimizes decision-making, significantly boosting diagnostic accuracy and clinical support. We also develop a reasoning data synthesis method, generating step-by-step reasoning data via rejection sampling, which further enhances the model's generalization. Experimental results show that after RL training, GMAI-VL-R1 excels in tasks such as medical image diagnosis and visual question answering. While the model demonstrates basic memorization with supervised fine-tuning, RL is crucial for true generalization. Our work establishes new evaluation benchmarks and paves the way for future advancements in medical reasoning models. Code, data, and model will be released at \href{https://github.com/uni-medical/GMAI-VL-R1}{this link}.
Towards Interpretable Counterfactual Generation via Multimodal Autoregression
Mar 29, 2025Abstract:Counterfactual medical image generation enables clinicians to explore clinical hypotheses, such as predicting disease progression, facilitating their decision-making. While existing methods can generate visually plausible images from disease progression prompts, they produce silent predictions that lack interpretation to verify how the generation reflects the hypothesized progression -- a critical gap for medical applications that require traceable reasoning. In this paper, we propose Interpretable Counterfactual Generation (ICG), a novel task requiring the joint generation of counterfactual images that reflect the clinical hypothesis and interpretation texts that outline the visual changes induced by the hypothesis. To enable ICG, we present ICG-CXR, the first dataset pairing longitudinal medical images with hypothetical progression prompts and textual interpretations. We further introduce ProgEmu, an autoregressive model that unifies the generation of counterfactual images and textual interpretations. We demonstrate the superiority of ProgEmu in generating progression-aligned counterfactuals and interpretations, showing significant potential in enhancing clinical decision support and medical education. Project page: https://progemu.github.io.
Radiologist-in-the-Loop Self-Training for Generalizable CT Metal Artifact Reduction
Jan 26, 2025



Abstract:Metal artifacts in computed tomography (CT) images can significantly degrade image quality and impede accurate diagnosis. Supervised metal artifact reduction (MAR) methods, trained using simulated datasets, often struggle to perform well on real clinical CT images due to a substantial domain gap. Although state-of-the-art semi-supervised methods use pseudo ground-truths generated by a prior network to mitigate this issue, their reliance on a fixed prior limits both the quality and quantity of these pseudo ground-truths, introducing confirmation bias and reducing clinical applicability. To address these limitations, we propose a novel Radiologist-In-the-loop SElf-training framework for MAR, termed RISE-MAR, which can integrate radiologists' feedback into the semi-supervised learning process, progressively improving the quality and quantity of pseudo ground-truths for enhanced generalization on real clinical CT images. For quality assurance, we introduce a clinical quality assessor model that emulates radiologist evaluations, effectively selecting high-quality pseudo ground-truths for semi-supervised training. For quantity assurance, our self-training framework iteratively generates additional high-quality pseudo ground-truths, expanding the clinical dataset and further improving model generalization. Extensive experimental results on multiple clinical datasets demonstrate the superior generalization performance of our RISE-MAR over state-of-the-art methods, advancing the development of MAR models for practical application. Code is available at https://github.com/Masaaki-75/rise-mar.
GMAI-VL & GMAI-VL-5.5M: A Large Vision-Language Model and A Comprehensive Multimodal Dataset Towards General Medical AI
Nov 21, 2024Abstract:Despite significant advancements in general artificial intelligence, such as GPT-4, their effectiveness in the medical domain (general medical AI, GMAI) remains constrained due to the absence of specialized medical knowledge. To address this challenge, we present GMAI-VL-5.5M, a comprehensive multimodal medical dataset created by converting hundreds of specialized medical datasets into meticulously constructed image-text pairs. This dataset features comprehensive task coverage, diverse modalities, and high-quality image-text data. Building upon this multimodal dataset, we propose GMAI-VL, a general medical vision-language model with a progressively three-stage training strategy. This approach significantly enhances the model's ability by integrating visual and textual information, thereby improving its ability to process multimodal data and support accurate diagnosis and clinical decision-making. Experimental evaluations demonstrate that GMAI-VL achieves state-of-the-art results across a wide range of multimodal medical tasks, such as visual question answering and medical image diagnosis. Our contributions include the development of the GMAI-VL-5.5M dataset, the introduction of the GMAI-VL model, and the establishment of new benchmarks in multiple medical domains. Code and dataset will be released at https://github.com/uni-medical/GMAI-VL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge