Ching-Wei Wang
*: shared first/last authors
Deep Learning Techniques for Automatic Lateral X-ray Cephalometric Landmark Detection: Is the Problem Solved?
Sep 24, 2024Abstract:Localization of the craniofacial landmarks from lateral cephalograms is a fundamental task in cephalometric analysis. The automation of the corresponding tasks has thus been the subject of intense research over the past decades. In this paper, we introduce the "Cephalometric Landmark Detection (CL-Detection)" dataset, which is the largest publicly available and comprehensive dataset for cephalometric landmark detection. This multi-center and multi-vendor dataset includes 600 lateral X-ray images with 38 landmarks acquired with different equipment from three medical centers. The overarching objective of this paper is to measure how far state-of-the-art deep learning methods can go for cephalometric landmark detection. Following the 2023 MICCAI CL-Detection Challenge, we report the results of the top ten research groups using deep learning methods. Results show that the best methods closely approximate the expert analysis, achieving a mean detection rate of 75.719% and a mean radial error of 1.518 mm. While there is room for improvement, these findings undeniably open the door to highly accurate and fully automatic location of craniofacial landmarks. We also identify scenarios for which deep learning methods are still failing. Both the dataset and detailed results are publicly available online, while the platform will remain open for the community to benchmark future algorithm developments at https://cl-detection2023.grand-challenge.org/.
Dual channel CW nnU-Net for 3D PET-CT Lesion Segmentation in 2024 autoPET III Challenge
Sep 11, 2024Abstract:PET/CT is extensively used in imaging malignant tumors because it highlights areas of increased glucose metabolism, indicative of cancerous activity. Accurate 3D lesion segmentation in PET/CT imaging is essential for effective oncological diagnostics and treatment planning. In this study, we developed an advanced 3D residual U-Net model for the Automated Lesion Segmentation in Whole-Body PET/CT - Multitracer Multicenter Generalization (autoPET III) Challenge, which will be held jointly with 2024 Medical Image Computing and Computer Assisted Intervention (MICCAI) conference at Marrakesh, Morocco. Proposed model incorporates a novel sample attention boosting technique to enhance segmentation performance by adjusting the contribution of challenging cases during training, improving generalization across FDG and PSMA tracers. The proposed model outperformed the challenge baseline model in the preliminary test set on the Grand Challenge platform, and our team is currently ranking in the 2nd place among 497 participants worldwide from 53 countries (accessed date: 2024/9/4), with Dice score of 0.8700, False Negative Volume of 19.3969 and False Positive Volume of 1.0857.
Is the winner really the best? A critical analysis of common research practice in biomedical image analysis competitions
Jun 06, 2018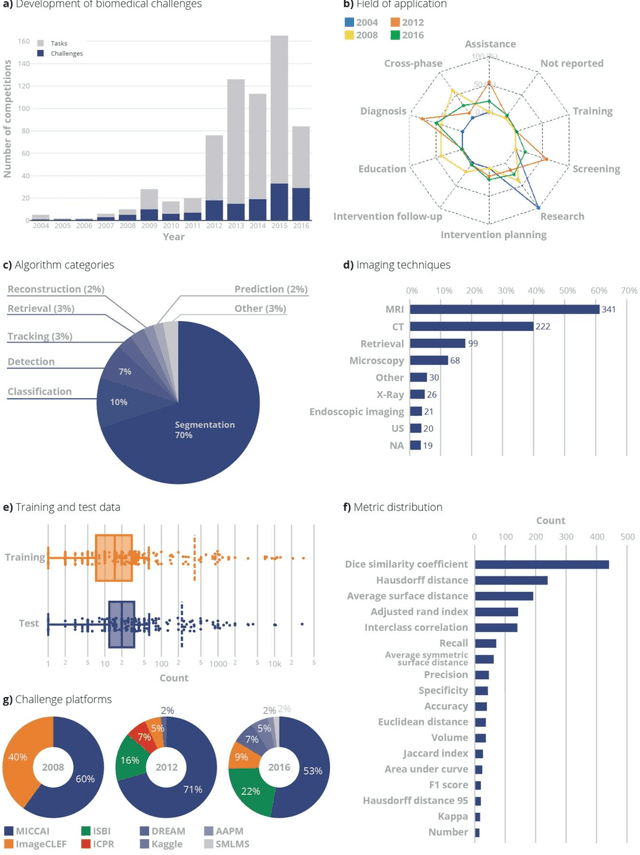
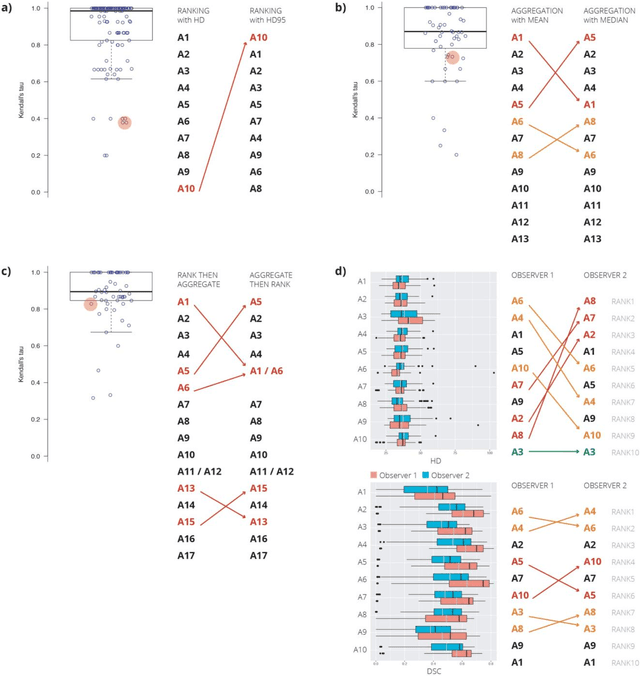
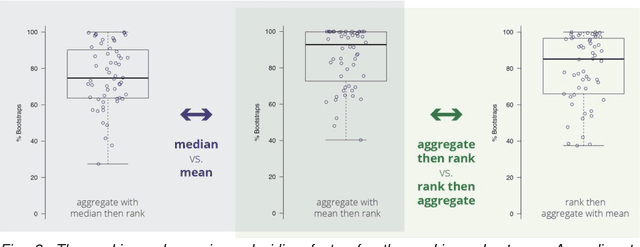
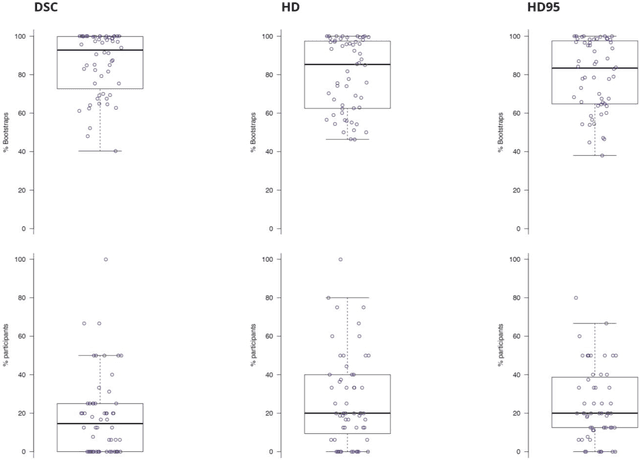
Abstract:International challenges have become the standard for validation of biomedical image analysis methods. Given their scientific impact, it is surprising that a critical analysis of common practices related to the organization of challenges has not yet been performed. In this paper, we present a comprehensive analysis of biomedical image analysis challenges conducted up to now. We demonstrate the importance of challenges and show that the lack of quality control has critical consequences. First, reproducibility and interpretation of the results is often hampered as only a fraction of relevant information is typically provided. Second, the rank of an algorithm is generally not robust to a number of variables such as the test data used for validation, the ranking scheme applied and the observers that make the reference annotations. To overcome these problems, we recommend best practice guidelines and define open research questions to be addressed in the future.
Assessment of algorithms for mitosis detection in breast cancer histopathology images
Nov 21, 2014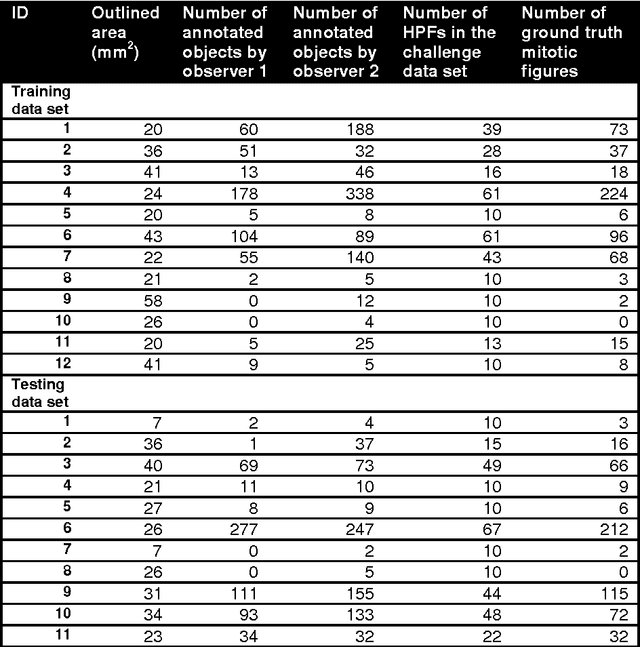
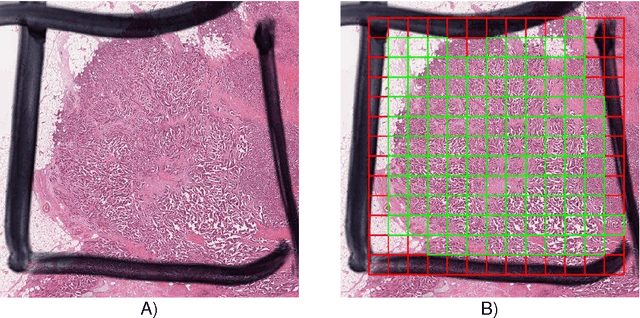
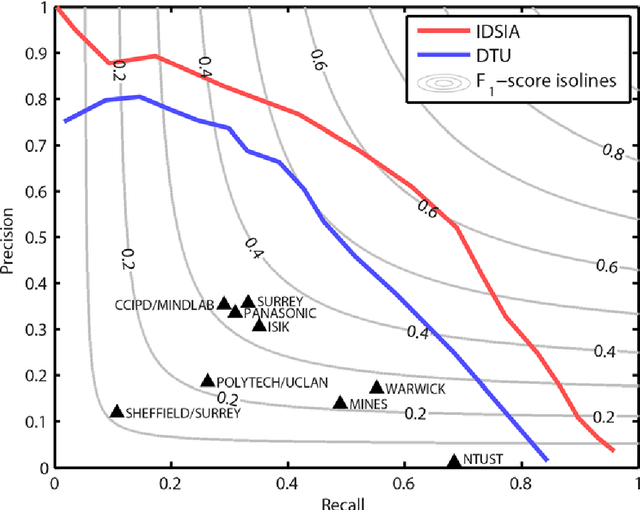
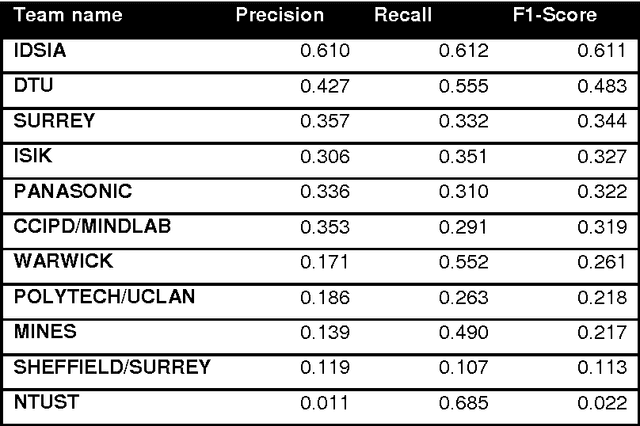
Abstract:The proliferative activity of breast tumors, which is routinely estimated by counting of mitotic figures in hematoxylin and eosin stained histology sections, is considered to be one of the most important prognostic markers. However, mitosis counting is laborious, subjective and may suffer from low inter-observer agreement. With the wider acceptance of whole slide images in pathology labs, automatic image analysis has been proposed as a potential solution for these issues. In this paper, the results from the Assessment of Mitosis Detection Algorithms 2013 (AMIDA13) challenge are described. The challenge was based on a data set consisting of 12 training and 11 testing subjects, with more than one thousand annotated mitotic figures by multiple observers. Short descriptions and results from the evaluation of eleven methods are presented. The top performing method has an error rate that is comparable to the inter-observer agreement among pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge