Aaron Carass
*: shared first/last authors
Likelihood-Separable Diffusion Inference for Multi-Image MRI Super-Resolution
Jan 20, 2026Abstract:Diffusion models are the current state-of-the-art for solving inverse problems in imaging. Their impressive generative capability allows them to approximate sampling from a prior distribution, which alongside a known likelihood function permits posterior sampling without retraining the model. While recent methods have made strides in advancing the accuracy of posterior sampling, the majority focuses on single-image inverse problems. However, for modalities such as magnetic resonance imaging (MRI), it is common to acquire multiple complementary measurements, each low-resolution along a different axis. In this work, we generalize common diffusion-based inverse single-image problem solvers for multi-image super-resolution (MISR) MRI. We show that the DPS likelihood correction allows an exactly-separable gradient decomposition across independently acquired measurements, enabling MISR without constructing a joint operator, modifying the diffusion model, or increasing network function evaluations. We derive MISR versions of DPS, DMAP, DPPS, and diffusion-based PnP/ADMM, and demonstrate substantial gains over SISR across $4\times/8\times/16\times$ anisotropic degradations. Our results achieve state-of-the-art super-resolution of anisotropic MRI volumes and, critically, enable reconstruction of near-isotropic anatomy from routine 2D multi-slice acquisitions, which are otherwise highly degraded in orthogonal views.
MSRepaint: Multiple Sclerosis Repaint with Conditional Denoising Diffusion Implicit Model for Bidirectional Lesion Filling and Synthesis
Oct 02, 2025Abstract:In multiple sclerosis, lesions interfere with automated magnetic resonance imaging analyses such as brain parcellation and deformable registration, while lesion segmentation models are hindered by the limited availability of annotated training data. To address both issues, we propose MSRepaint, a unified diffusion-based generative model for bidirectional lesion filling and synthesis that restores anatomical continuity for downstream analyses and augments segmentation through realistic data generation. MSRepaint conditions on spatial lesion masks for voxel-level control, incorporates contrast dropout to handle missing inputs, integrates a repainting mechanism to preserve surrounding anatomy during lesion filling and synthesis, and employs a multi-view DDIM inversion and fusion pipeline for 3D consistency with fast inference. Extensive evaluations demonstrate the effectiveness of MSRepaint across multiple tasks. For lesion filling, we evaluate both the accuracy within the filled regions and the impact on downstream tasks including brain parcellation and deformable registration. MSRepaint outperforms the traditional lesion filling methods FSL and NiftySeg, and achieves accuracy on par with FastSurfer-LIT, a recent diffusion model-based inpainting method, while offering over 20 times faster inference. For lesion synthesis, state-of-the-art MS lesion segmentation models trained on MSRepaint-synthesized data outperform those trained on CarveMix-synthesized data or real ISBI challenge training data across multiple benchmarks, including the MICCAI 2016 and UMCL datasets. Additionally, we demonstrate that MSRepaint's unified bidirectional filling and synthesis capability, with full spatial control over lesion appearance, enables high-fidelity simulation of lesion evolution in longitudinal MS progression.
UNISELF: A Unified Network with Instance Normalization and Self-Ensembled Lesion Fusion for Multiple Sclerosis Lesion Segmentation
Aug 06, 2025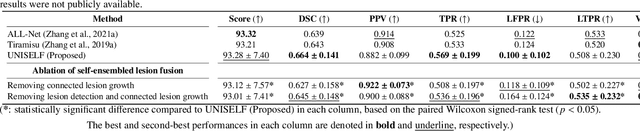
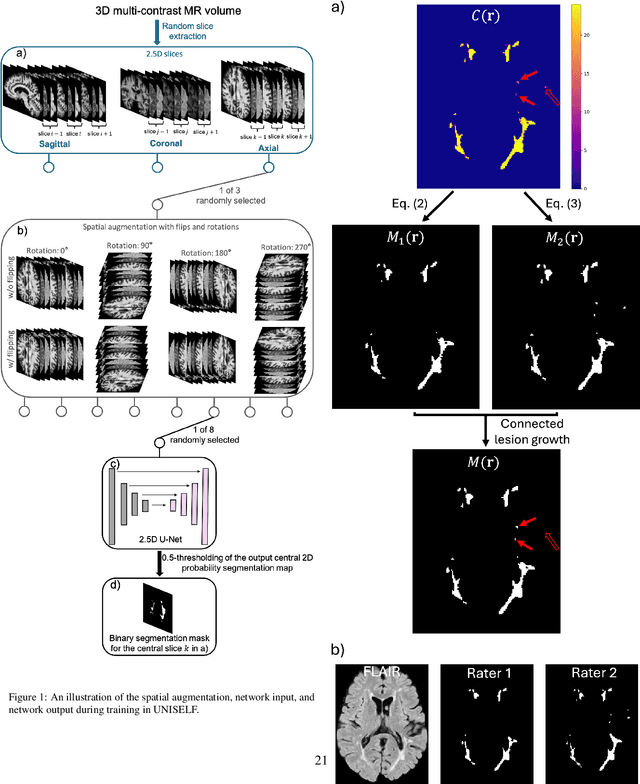
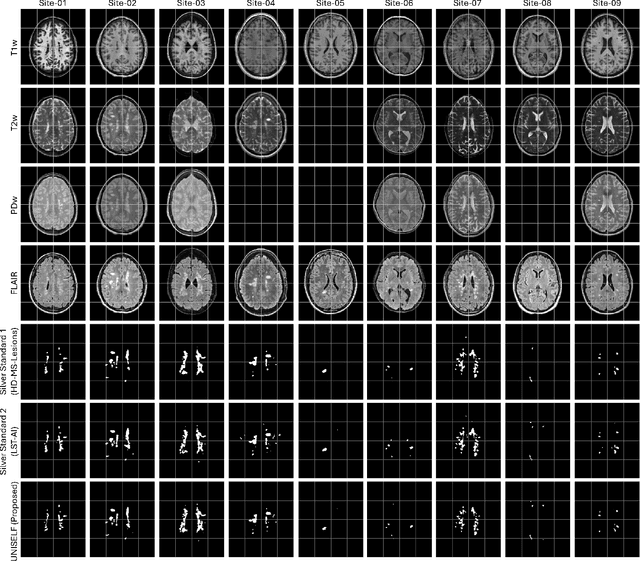

Abstract:Automated segmentation of multiple sclerosis (MS) lesions using multicontrast magnetic resonance (MR) images improves efficiency and reproducibility compared to manual delineation, with deep learning (DL) methods achieving state-of-the-art performance. However, these DL-based methods have yet to simultaneously optimize in-domain accuracy and out-of-domain generalization when trained on a single source with limited data, or their performance has been unsatisfactory. To fill this gap, we propose a method called UNISELF, which achieves high accuracy within a single training domain while demonstrating strong generalizability across multiple out-of-domain test datasets. UNISELF employs a novel test-time self-ensembled lesion fusion to improve segmentation accuracy, and leverages test-time instance normalization (TTIN) of latent features to address domain shifts and missing input contrasts. Trained on the ISBI 2015 longitudinal MS segmentation challenge training dataset, UNISELF ranks among the best-performing methods on the challenge test dataset. Additionally, UNISELF outperforms all benchmark methods trained on the same ISBI training data across diverse out-of-domain test datasets with domain shifts and missing contrasts, including the public MICCAI 2016 and UMCL datasets, as well as a private multisite dataset. These test datasets exhibit domain shifts and/or missing contrasts caused by variations in acquisition protocols, scanner types, and imaging artifacts arising from imperfect acquisition. Our code is available at https://github.com/uponacceptance.
Synthetic multi-inversion time magnetic resonance images for visualization of subcortical structures
Jun 04, 2025Abstract:Purpose: Visualization of subcortical gray matter is essential in neuroscience and clinical practice, particularly for disease understanding and surgical planning.While multi-inversion time (multi-TI) T$_1$-weighted (T$_1$-w) magnetic resonance (MR) imaging improves visualization, it is rarely acquired in clinical settings. Approach: We present SyMTIC (Synthetic Multi-TI Contrasts), a deep learning method that generates synthetic multi-TI images using routinely acquired T$_1$-w, T$_2$-weighted (T$_2$-w), and FLAIR images. Our approach combines image translation via deep neural networks with imaging physics to estimate longitudinal relaxation time (T$_1$) and proton density (PD) maps. These maps are then used to compute multi-TI images with arbitrary inversion times. Results: SyMTIC was trained using paired MPRAGE and FGATIR images along with T$_2$-w and FLAIR images. It accurately synthesized multi-TI images from standard clinical inputs, achieving image quality comparable to that from explicitly acquired multi-TI data.The synthetic images, especially for TI values between 400-800 ms, enhanced visualization of subcortical structures and improved segmentation of thalamic nuclei. Conclusion: SyMTIC enables robust generation of high-quality multi-TI images from routine MR contrasts. It generalizes well to varied clinical datasets, including those with missing FLAIR images or unknown parameters, offering a practical solution for improving brain MR image visualization and analysis.
Pretraining Deformable Image Registration Networks with Random Images
May 30, 2025Abstract:Recent advances in deep learning-based medical image registration have shown that training deep neural networks~(DNNs) does not necessarily require medical images. Previous work showed that DNNs trained on randomly generated images with carefully designed noise and contrast properties can still generalize well to unseen medical data. Building on this insight, we propose using registration between random images as a proxy task for pretraining a foundation model for image registration. Empirical results show that our pretraining strategy improves registration accuracy, reduces the amount of domain-specific data needed to achieve competitive performance, and accelerates convergence during downstream training, thereby enhancing computational efficiency.
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025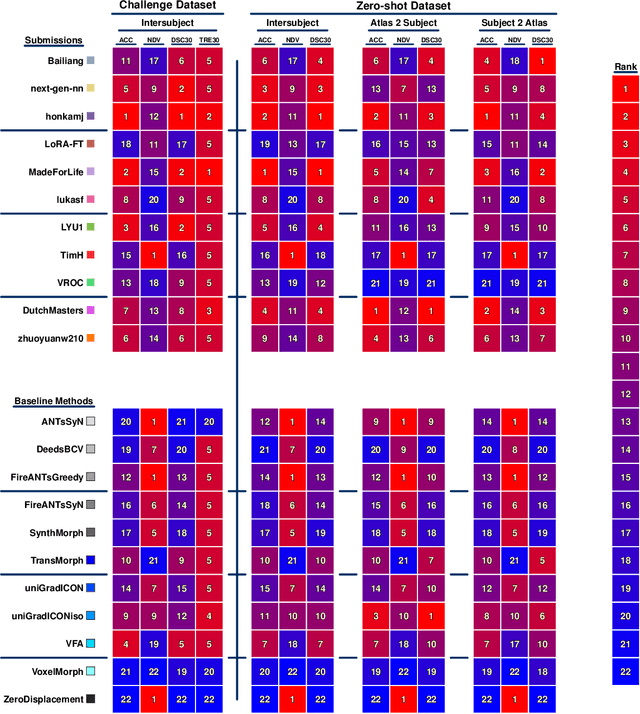
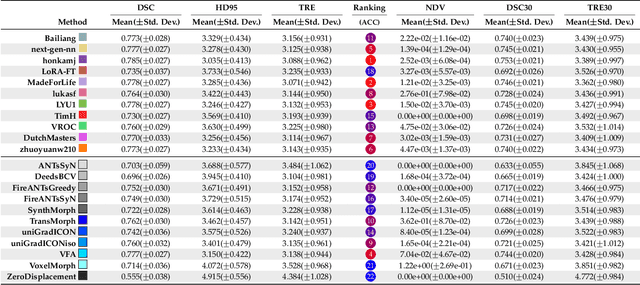
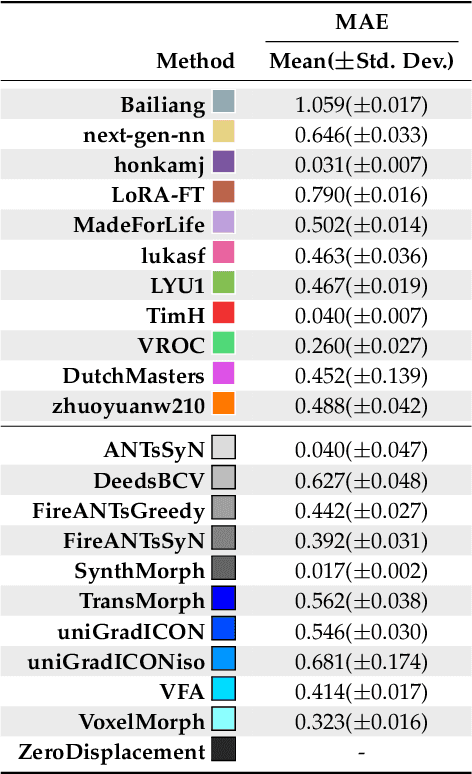
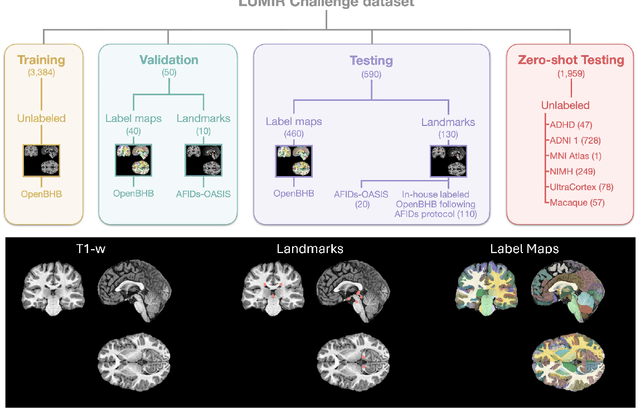
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
Brightness-Invariant Tracking Estimation in Tagged MRI
May 23, 2025Abstract:Magnetic resonance (MR) tagging is an imaging technique for noninvasively tracking tissue motion in vivo by creating a visible pattern of magnetization saturation (tags) that deforms with the tissue. Due to longitudinal relaxation and progression to steady-state, the tags and tissue brightnesses change over time, which makes tracking with optical flow methods error-prone. Although Fourier methods can alleviate these problems, they are also sensitive to brightness changes as well as spectral spreading due to motion. To address these problems, we introduce the brightness-invariant tracking estimation (BRITE) technique for tagged MRI. BRITE disentangles the anatomy from the tag pattern in the observed tagged image sequence and simultaneously estimates the Lagrangian motion. The inherent ill-posedness of this problem is addressed by leveraging the expressive power of denoising diffusion probabilistic models to represent the probabilistic distribution of the underlying anatomy and the flexibility of physics-informed neural networks to estimate biologically-plausible motion. A set of tagged MR images of a gel phantom was acquired with various tag periods and imaging flip angles to demonstrate the impact of brightness variations and to validate our method. The results show that BRITE achieves more accurate motion and strain estimates as compared to other state of the art methods, while also being resistant to tag fading.
Correlation Ratio for Unsupervised Learning of Multi-modal Deformable Registration
Apr 16, 2025Abstract:In recent years, unsupervised learning for deformable image registration has been a major research focus. This approach involves training a registration network using pairs of moving and fixed images, along with a loss function that combines an image similarity measure and deformation regularization. For multi-modal image registration tasks, the correlation ratio has been a widely-used image similarity measure historically, yet it has been underexplored in current deep learning methods. Here, we propose a differentiable correlation ratio to use as a loss function for learning-based multi-modal deformable image registration. This approach extends the traditionally non-differentiable implementation of the correlation ratio by using the Parzen windowing approximation, enabling backpropagation with deep neural networks. We validated the proposed correlation ratio on a multi-modal neuroimaging dataset. In addition, we established a Bayesian training framework to study how the trade-off between the deformation regularizer and similarity measures, including mutual information and our proposed correlation ratio, affects the registration performance.
ECLARE: Efficient cross-planar learning for anisotropic resolution enhancement
Mar 14, 2025Abstract:In clinical imaging, magnetic resonance (MR) image volumes are often acquired as stacks of 2D slices, permitting decreased scan times, improved signal-to-noise ratio, and image contrasts unique to 2D MR pulse sequences. While this is sufficient for clinical evaluation, automated algorithms designed for 3D analysis perform sub-optimally on 2D-acquired scans, especially those with thick slices and gaps between slices. Super-resolution (SR) methods aim to address this problem, but previous methods do not address all of the following: slice profile shape estimation, slice gap, domain shift, and non-integer / arbitrary upsampling factors. In this paper, we propose ECLARE (Efficient Cross-planar Learning for Anisotropic Resolution Enhancement), a self-SR method that addresses each of these factors. ECLARE estimates the slice profile from the 2D-acquired multi-slice MR volume, trains a network to learn the mapping from low-resolution to high-resolution in-plane patches from the same volume, and performs SR with anti-aliasing. We compared ECLARE to cubic B-spline interpolation, SMORE, and other contemporary SR methods. We used realistic and representative simulations so that quantitative performance against a ground truth could be computed, and ECLARE outperformed all other methods in both signal recovery and downstream tasks. On real data for which there is no ground truth, ECLARE demonstrated qualitative superiority over other methods as well. Importantly, as ECLARE does not use external training data it cannot suffer from domain shift between training and testing. Our code is open-source and available at https://www.github.com/sremedios/eclare.
Unsupervised learning of spatially varying regularization for diffeomorphic image registration
Dec 23, 2024Abstract:Spatially varying regularization accommodates the deformation variations that may be necessary for different anatomical regions during deformable image registration. Historically, optimization-based registration models have harnessed spatially varying regularization to address anatomical subtleties. However, most modern deep learning-based models tend to gravitate towards spatially invariant regularization, wherein a homogenous regularization strength is applied across the entire image, potentially disregarding localized variations. In this paper, we propose a hierarchical probabilistic model that integrates a prior distribution on the deformation regularization strength, enabling the end-to-end learning of a spatially varying deformation regularizer directly from the data. The proposed method is straightforward to implement and easily integrates with various registration network architectures. Additionally, automatic tuning of hyperparameters is achieved through Bayesian optimization, allowing efficient identification of optimal hyperparameters for any given registration task. Comprehensive evaluations on publicly available datasets demonstrate that the proposed method significantly improves registration performance and enhances the interpretability of deep learning-based registration, all while maintaining smooth deformations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge