Yihao Liu
Training-Free Acceleration for Document Parsing Vision-Language Model with Hierarchical Speculative Decoding
Feb 13, 2026Abstract:Document parsing is a fundamental task in multimodal understanding, supporting a wide range of downstream applications such as information extraction and intelligent document analysis. Benefiting from strong semantic modeling and robust generalization, VLM-based end-to-end approaches have emerged as the mainstream paradigm in recent years. However, these models often suffer from substantial inference latency, as they must auto-regressively generate long token sequences when processing long-form documents. In this work, motivated by the extremely long outputs and complex layout structures commonly found in document parsing, we propose a training-free and highly efficient acceleration method. Inspired by speculative decoding, we employ a lightweight document parsing pipeline as a draft model to predict batches of future tokens, while the more accurate VLM verifies these draft predictions in parallel. Moreover, we further exploit the layout-structured nature of documents by partitioning each page into independent regions, enabling parallel decoding of each region using the same draft-verify strategy. The final predictions are then assembled according to the natural reading order. Experimental results demonstrate the effectiveness of our approach: on the general-purpose OmniDocBench, our method provides a 2.42x lossless acceleration for the dots.ocr model, and achieves up to 4.89x acceleration on long-document parsing tasks. We will release our code to facilitate reproducibility and future research.
FlowMind: Execute-Summarize for Structured Workflow Generation from LLM Reasoning
Feb 12, 2026Abstract:LLMs can solve complex tasks through reasoning and tool use, but accurately translating these solutions into structured workflows remains challenging. We model workflows as sequences of tool use and reformulate the problem as designing a mechanism that can both solve tasks and reliably construct workflows. Prior approaches that build workflows during execution often suffer from inaccuracies due to interference between the two processes. We propose an Execute-Summarize(ES) framework that decouples task execution from workflow construction: the model first completes the task using available tools, then independently reconstructs a structured workflow from execution traces. This separation improves workflow accuracy and robustness. We introduce FlowBench and show through extensive experiments that our approach outperforms existing methods, providing a reliable paradigm for grounding free-form LLM reasoning into structured workflows.
InternAgent-1.5: A Unified Agentic Framework for Long-Horizon Autonomous Scientific Discovery
Feb 09, 2026Abstract:We introduce InternAgent-1.5, a unified system designed for end-to-end scientific discovery across computational and empirical domains. The system is built on a structured architecture composed of three coordinated subsystems for generation, verification, and evolution. These subsystems are supported by foundational capabilities for deep research, solution optimization, and long horizon memory. The architecture allows InternAgent-1.5 to operate continuously across extended discovery cycles while maintaining coherent and improving behavior. It also enables the system to coordinate computational modeling and laboratory experimentation within a single unified system. We evaluate InternAgent-1.5 on scientific reasoning benchmarks such as GAIA, HLE, GPQA, and FrontierScience, and the system achieves leading performance that demonstrates strong foundational capabilities. Beyond these benchmarks, we further assess two categories of discovery tasks. In algorithm discovery tasks, InternAgent-1.5 autonomously designs competitive methods for core machine learning problems. In empirical discovery tasks, it executes complete computational or wet lab experiments and produces scientific findings in earth, life, biological, and physical domains. Overall, these results show that InternAgent-1.5 provides a general and scalable framework for autonomous scientific discovery.
SupChain-Bench: Benchmarking Large Language Models for Real-World Supply Chain Management
Feb 07, 2026Abstract:Large language models (LLMs) have shown promise in complex reasoning and tool-based decision making, motivating their application to real-world supply chain management. However, supply chain workflows require reliable long-horizon, multi-step orchestration grounded in domain-specific procedures, which remains challenging for current models. To systematically evaluate LLM performance in this setting, we introduce SupChain-Bench, a unified real-world benchmark that assesses both supply chain domain knowledge and long-horizon tool-based orchestration grounded in standard operating procedures (SOPs). Our experiments reveal substantial gaps in execution reliability across models. We further propose SupChain-ReAct, an SOP-free framework that autonomously synthesizes executable procedures for tool use, achieving the strongest and most consistent tool-calling performance. Our work establishes a principled benchmark for studying reliable long-horizon orchestration in real-world operational settings and highlights significant room for improvement in LLM-based supply chain agents.
Toward Generalizable Deblurring: Leveraging Massive Blur Priors with Linear Attention for Real-World Scenarios
Jan 10, 2026Abstract:Image deblurring has advanced rapidly with deep learning, yet most methods exhibit poor generalization beyond their training datasets, with performance dropping significantly in real-world scenarios. Our analysis shows this limitation stems from two factors: datasets face an inherent trade-off between realism and coverage of diverse blur patterns, and algorithmic designs remain restrictive, as pixel-wise losses drive models toward local detail recovery while overlooking structural and semantic consistency, whereas diffusion-based approaches, though perceptually strong, still fail to generalize when trained on narrow datasets with simplistic strategies. Through systematic investigation, we identify blur pattern diversity as the decisive factor for robust generalization and propose Blur Pattern Pretraining (BPP), which acquires blur priors from simulation datasets and transfers them through joint fine-tuning on real data. We further introduce Motion and Semantic Guidance (MoSeG) to strengthen blur priors under severe degradation, and integrate it into GLOWDeblur, a Generalizable reaL-wOrld lightWeight Deblur model that combines convolution-based pre-reconstruction & domain alignment module with a lightweight diffusion backbone. Extensive experiments on six widely-used benchmarks and two real-world datasets validate our approach, confirming the importance of blur priors for robust generalization and demonstrating that the lightweight design of GLOWDeblur ensures practicality in real-world applications. The project page is available at https://vegdog007.github.io/GLOWDeblur_Website/.
UniPercept: Towards Unified Perceptual-Level Image Understanding across Aesthetics, Quality, Structure, and Texture
Dec 25, 2025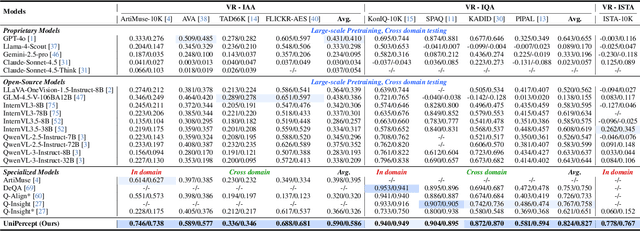
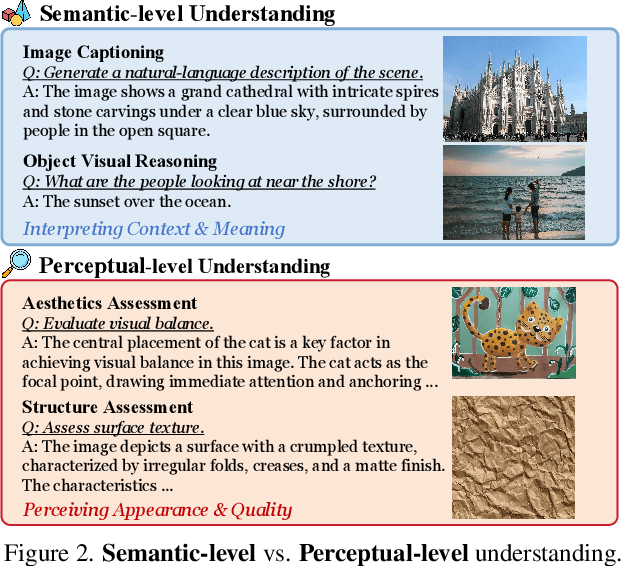
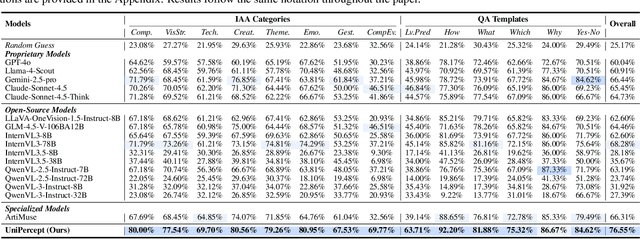
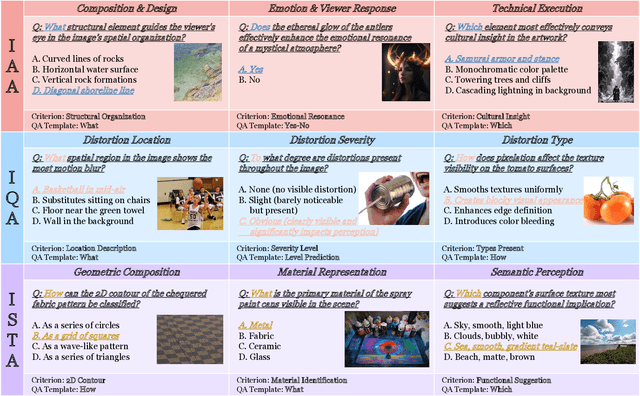
Abstract:Multimodal large language models (MLLMs) have achieved remarkable progress in visual understanding tasks such as visual grounding, segmentation, and captioning. However, their ability to perceive perceptual-level image features remains limited. In this work, we present UniPercept-Bench, a unified framework for perceptual-level image understanding across three key domains: Aesthetics, Quality, Structure and Texture. We establish a hierarchical definition system and construct large-scale datasets to evaluate perceptual-level image understanding. Based on this foundation, we develop a strong baseline UniPercept trained via Domain-Adaptive Pre-Training and Task-Aligned RL, enabling robust generalization across both Visual Rating (VR) and Visual Question Answering (VQA) tasks. UniPercept outperforms existing MLLMs on perceptual-level image understanding and can serve as a plug-and-play reward model for text-to-image generation. This work defines Perceptual-Level Image Understanding in the era of MLLMs and, through the introduction of a comprehensive benchmark together with a strong baseline, provides a solid foundation for advancing perceptual-level multimodal image understanding.
Omni-Weather: Unified Multimodal Foundation Model for Weather Generation and Understanding
Dec 25, 2025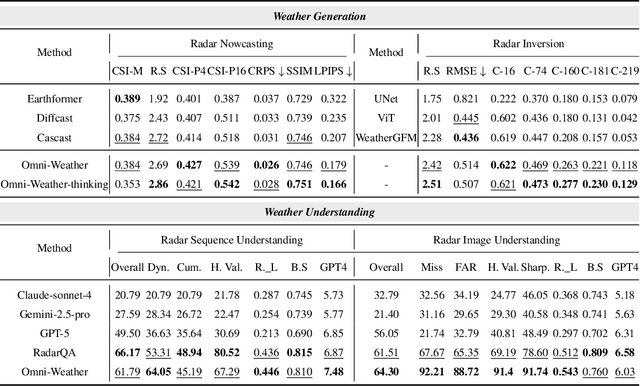
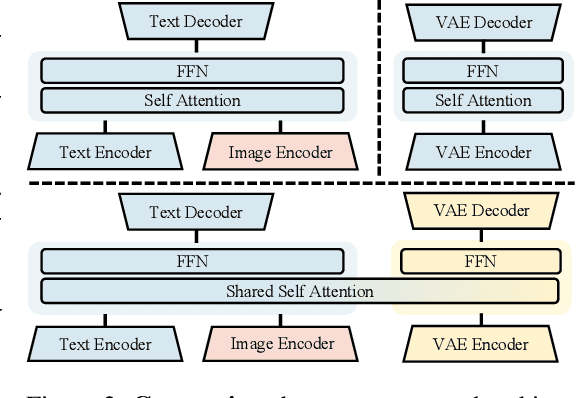
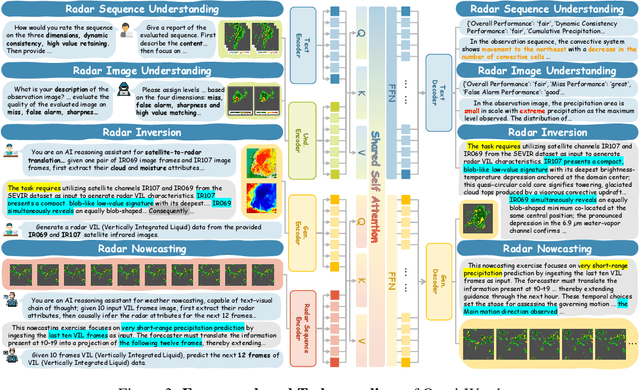
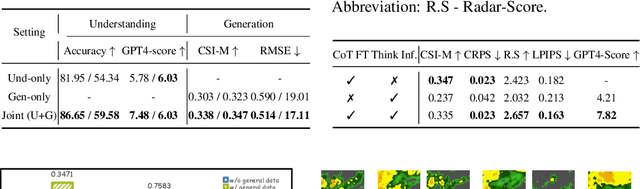
Abstract:Weather modeling requires both accurate prediction and mechanistic interpretation, yet existing methods treat these goals in isolation, separating generation from understanding. To address this gap, we present Omni-Weather, the first multimodal foundation model that unifies weather generation and understanding within a single architecture. Omni-Weather integrates a radar encoder for weather generation tasks, followed by unified processing using a shared self-attention mechanism. Moreover, we construct a Chain-of-Thought dataset for causal reasoning in weather generation, enabling interpretable outputs and improved perceptual quality. Extensive experiments show Omni-Weather achieves state-of-the-art performance in both weather generation and understanding. Our findings further indicate that generative and understanding tasks in the weather domain can mutually enhance each other. Omni-Weather also demonstrates the feasibility and value of unifying weather generation and understanding.
dMLLM-TTS: Self-Verified and Efficient Test-Time Scaling for Diffusion Multi-Modal Large Language Models
Dec 22, 2025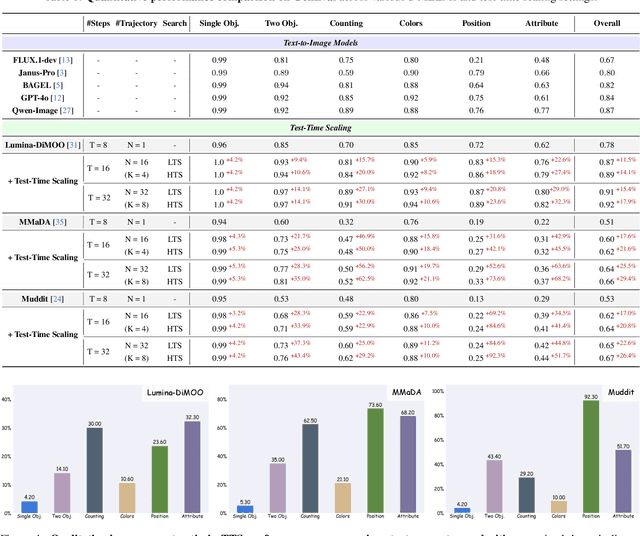
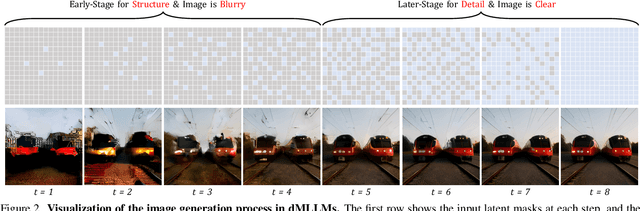
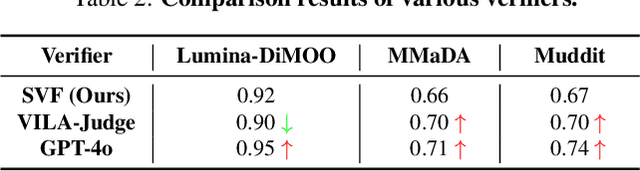
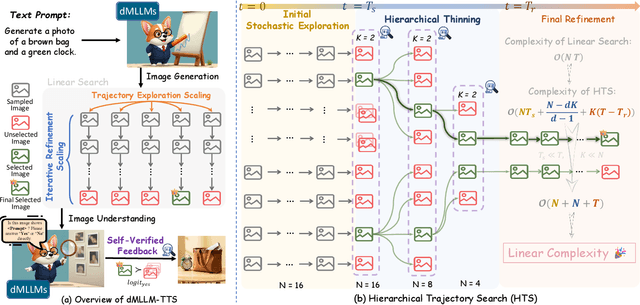
Abstract:Diffusion Multi-modal Large Language Models (dMLLMs) have recently emerged as a novel architecture unifying image generation and understanding. However, developing effective and efficient Test-Time Scaling (TTS) methods to unlock their full generative potential remains an underexplored challenge. To address this, we propose dMLLM-TTS, a novel framework operating on two complementary scaling axes: (1) trajectory exploration scaling to enhance the diversity of generated hypotheses, and (2) iterative refinement scaling for stable generation. Conventional TTS approaches typically perform linear search across these two dimensions, incurring substantial computational costs of O(NT) and requiring an external verifier for best-of-N selection. To overcome these limitations, we propose two innovations. First, we design an efficient hierarchical search algorithm with O(N+T) complexity that adaptively expands and prunes sampling trajectories. Second, we introduce a self-verified feedback mechanism that leverages the dMLLMs' intrinsic image understanding capabilities to assess text-image alignment, eliminating the need for external verifier. Extensive experiments on the GenEval benchmark across three representative dMLLMs (e.g., Lumina-DiMOO, MMaDA, Muddit) show that our framework substantially improves generation quality while achieving up to 6x greater efficiency than linear search. Project page: https://github.com/Alpha-VLLM/Lumina-DiMOO.
MetaVoxel: Joint Diffusion Modeling of Imaging and Clinical Metadata
Dec 12, 2025Abstract:Modern deep learning methods have achieved impressive results across tasks from disease classification, estimating continuous biomarkers, to generating realistic medical images. Most of these approaches are trained to model conditional distributions defined by a specific predictive direction with a specific set of input variables. We introduce MetaVoxel, a generative joint diffusion modeling framework that models the joint distribution over imaging data and clinical metadata by learning a single diffusion process spanning all variables. By capturing the joint distribution, MetaVoxel unifies tasks that traditionally require separate conditional models and supports flexible zero-shot inference using arbitrary subsets of inputs without task-specific retraining. Using more than 10,000 T1-weighted MRI scans paired with clinical metadata from nine datasets, we show that a single MetaVoxel model can perform image generation, age estimation, and sex prediction, achieving performance comparable to established task-specific baselines. Additional experiments highlight its capabilities for flexible inference. Together, these findings demonstrate that joint multimodal diffusion offers a promising direction for unifying medical AI models and enabling broader clinical applicability.
SynWeather: Weather Observation Data Synthesis across Multiple Regions and Variables via a General Diffusion Transformer
Nov 15, 2025Abstract:With the advancement of meteorological instruments, abundant data has become available. Current approaches are typically focus on single-variable, single-region tasks and primarily rely on deterministic modeling. This limits unified synthesis across variables and regions, overlooks cross-variable complementarity and often leads to over-smoothed results. To address above challenges, we introduce SynWeather, the first dataset designed for Unified Multi-region and Multi-variable Weather Observation Data Synthesis. SynWeather covers four representative regions: the Continental United States, Europe, East Asia, and Tropical Cyclone regions, as well as provides high-resolution observations of key weather variables, including Composite Radar Reflectivity, Hourly Precipitation, Visible Light, and Microwave Brightness Temperature. In addition, we introduce SynWeatherDiff, a general and probabilistic weather synthesis model built upon the Diffusion Transformer framework to address the over-smoothed problem. Experiments on the SynWeather dataset demonstrate the effectiveness of our network compared with both task-specific and general models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge