Junyu Chen
BPDQ: Bit-Plane Decomposition Quantization on a Variable Grid for Large Language Models
Feb 04, 2026Abstract:Large language model (LLM) inference is often bounded by memory footprint and memory bandwidth in resource-constrained deployments, making quantization a fundamental technique for efficient serving. While post-training quantization (PTQ) maintains high fidelity at 4-bit, it deteriorates at 2-3 bits. Fundamentally, existing methods enforce a shape-invariant quantization grid (e.g., the fixed uniform intervals of UINT2) for each group, severely restricting the feasible set for error minimization. To address this, we propose Bit-Plane Decomposition Quantization (BPDQ), which constructs a variable quantization grid via bit-planes and scalar coefficients, and iteratively refines them using approximate second-order information while progressively compensating quantization errors to minimize output discrepancy. In the 2-bit regime, BPDQ enables serving Qwen2.5-72B on a single RTX 3090 with 83.85% GSM8K accuracy (vs. 90.83% at 16-bit). Moreover, we provide theoretical analysis showing that the variable grid expands the feasible set, and that the quantization process consistently aligns with the optimization objective in Hessian-induced geometry. Code: github.com/KingdalfGoodman/BPDQ.
Deep classifier kriging for probabilistic spatial prediction of air quality index
Dec 29, 2025Abstract:Accurate spatial interpolation of the air quality index (AQI), computed from concentrations of multiple air pollutants, is essential for regulatory decision-making, yet AQI fields are inherently non-Gaussian and often exhibit complex nonlinear spatial structure. Classical spatial prediction methods such as kriging are linear and rely on Gaussian assumptions, which limits their ability to capture these features and to provide reliable predictive distributions. In this study, we propose \textit{deep classifier kriging} (DCK), a flexible, distribution-free deep learning framework for estimating full predictive distribution functions for univariate and bivariate spatial processes, together with a \textit{data fusion} mechanism that enables modeling of non-collocated bivariate processes and integration of heterogeneous air pollution data sources. Through extensive simulation experiments, we show that DCK consistently outperforms conventional approaches in predictive accuracy and uncertainty quantification. We further apply DCK to probabilistic spatial prediction of AQI by fusing sparse but high-quality station observations with spatially continuous yet biased auxiliary model outputs, yielding spatially resolved predictive distributions that support downstream tasks such as exceedance and extreme-event probability estimation for regulatory risk assessment and policy formulation.
Retrieval Augmented Generation-Enhanced Distributed LLM Agents for Generalizable Traffic Signal Control with Emergency Vehicles
Oct 30, 2025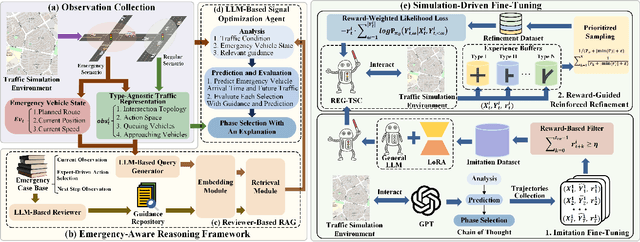
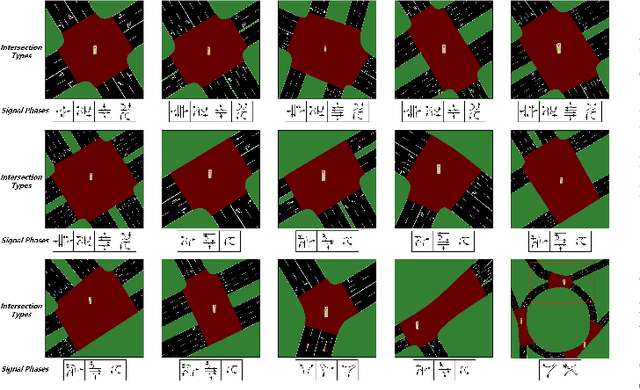
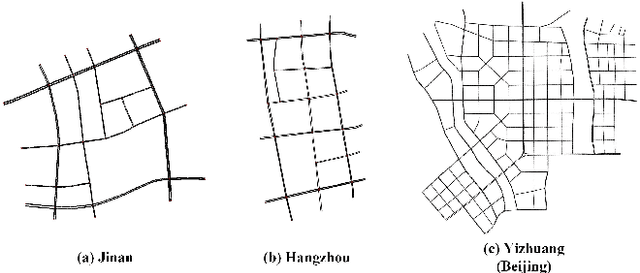
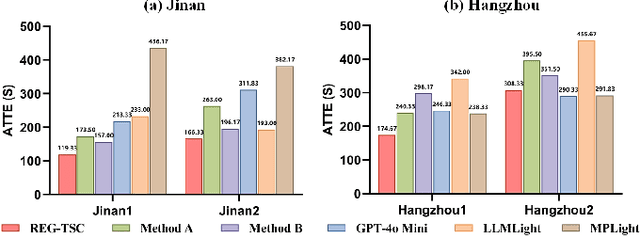
Abstract:With increasing urban traffic complexity, Traffic Signal Control (TSC) is essential for optimizing traffic flow and improving road safety. Large Language Models (LLMs) emerge as promising approaches for TSC. However, they are prone to hallucinations in emergencies, leading to unreliable decisions that may cause substantial delays for emergency vehicles. Moreover, diverse intersection types present substantial challenges for traffic state encoding and cross-intersection training, limiting generalization across heterogeneous intersections. Therefore, this paper proposes Retrieval Augmented Generation (RAG)-enhanced distributed LLM agents with Emergency response for Generalizable TSC (REG-TSC). Firstly, this paper presents an emergency-aware reasoning framework, which dynamically adjusts reasoning depth based on the emergency scenario and is equipped with a novel Reviewer-based Emergency RAG (RERAG) to distill specific knowledge and guidance from historical cases, enhancing the reliability and rationality of agents' emergency decisions. Secondly, this paper designs a type-agnostic traffic representation and proposes a Reward-guided Reinforced Refinement (R3) for heterogeneous intersections. R3 adaptively samples training experience from diverse intersections with environment feedback-based priority and fine-tunes LLM agents with a designed reward-weighted likelihood loss, guiding REG-TSC toward high-reward policies across heterogeneous intersections. On three real-world road networks with 17 to 177 heterogeneous intersections, extensive experiments show that REG-TSC reduces travel time by 42.00%, queue length by 62.31%, and emergency vehicle waiting time by 83.16%, outperforming other state-of-the-art methods.
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025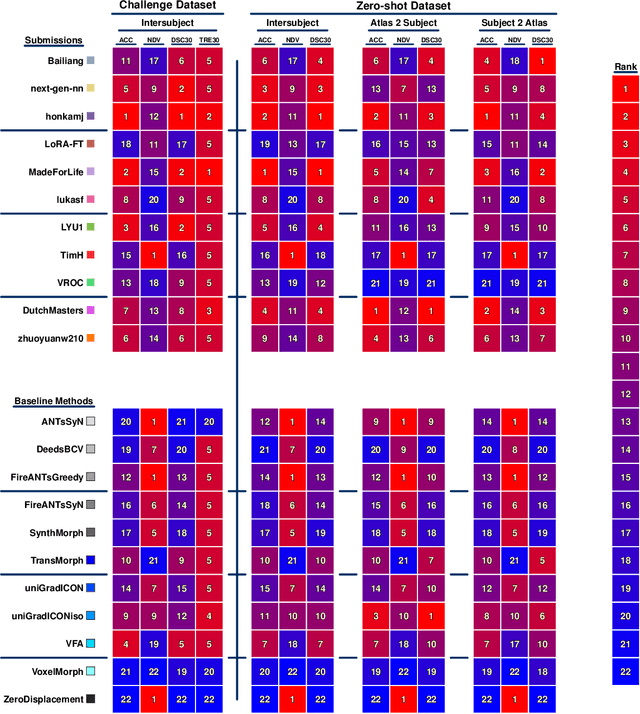
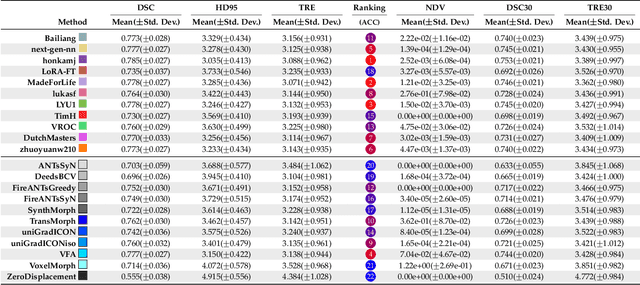
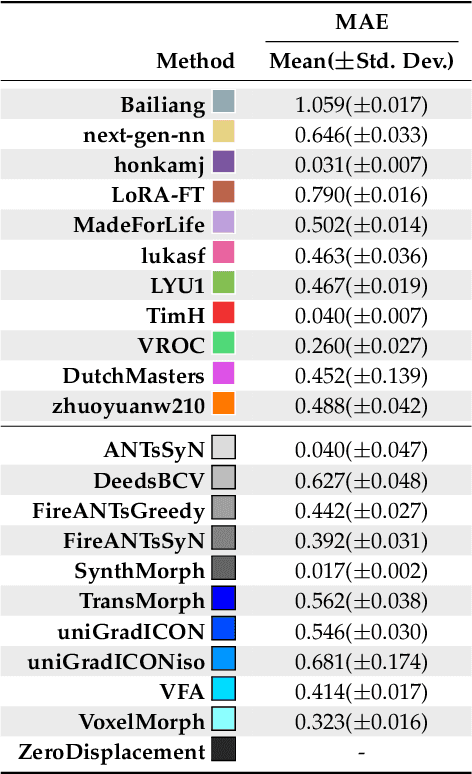
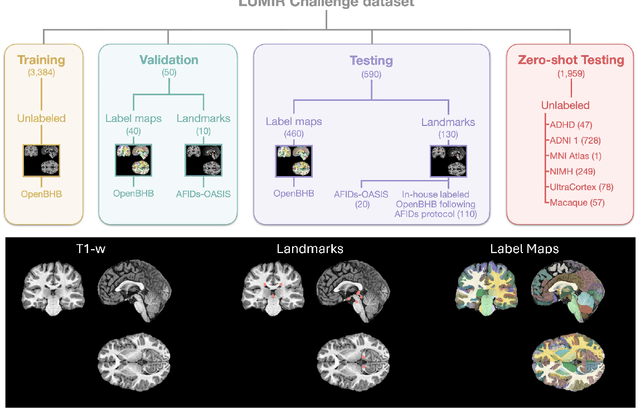
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
Pretraining Deformable Image Registration Networks with Random Images
May 30, 2025Abstract:Recent advances in deep learning-based medical image registration have shown that training deep neural networks~(DNNs) does not necessarily require medical images. Previous work showed that DNNs trained on randomly generated images with carefully designed noise and contrast properties can still generalize well to unseen medical data. Building on this insight, we propose using registration between random images as a proxy task for pretraining a foundation model for image registration. Empirical results show that our pretraining strategy improves registration accuracy, reduces the amount of domain-specific data needed to achieve competitive performance, and accelerates convergence during downstream training, thereby enhancing computational efficiency.
Deep learning-derived arterial input function
May 30, 2025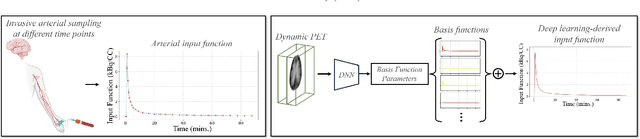
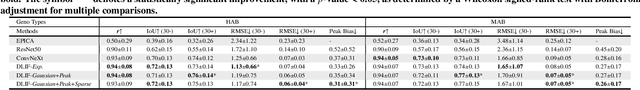
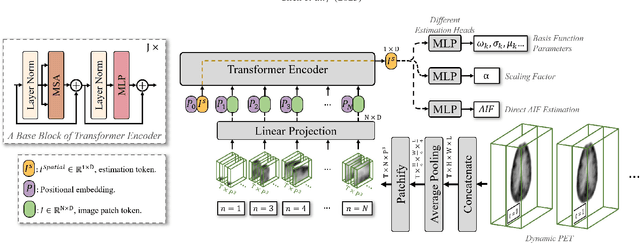
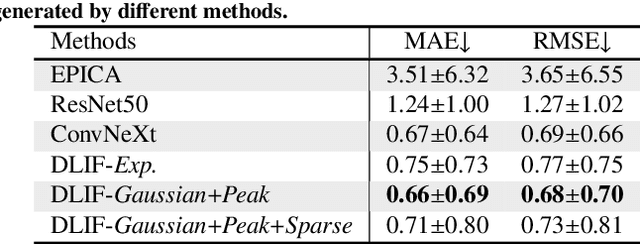
Abstract:Dynamic positron emission tomography (PET) imaging combined with radiotracer kinetic modeling is a powerful technique for visualizing biological processes in the brain, offering valuable insights into brain functions and neurological disorders such as Alzheimer's and Parkinson's diseases. Accurate kinetic modeling relies heavily on the use of a metabolite-corrected arterial input function (AIF), which typically requires invasive and labor-intensive arterial blood sampling. While alternative non-invasive approaches have been proposed, they often compromise accuracy or still necessitate at least one invasive blood sampling. In this study, we present the deep learning-derived arterial input function (DLIF), a deep learning framework capable of estimating a metabolite-corrected AIF directly from dynamic PET image sequences without any blood sampling. We validated DLIF using existing dynamic PET patient data. We compared DLIF and resulting parametric maps against ground truth measurements. Our evaluation shows that DLIF achieves accurate and robust AIF estimation. By leveraging deep learning's ability to capture complex temporal dynamics and incorporating prior knowledge of typical AIF shapes through basis functions, DLIF provides a rapid, accurate, and entirely non-invasive alternative to traditional AIF measurement methods.
LoTA-QAF: Lossless Ternary Adaptation for Quantization-Aware Fine-Tuning
May 24, 2025Abstract:Quantization and fine-tuning are crucial for deploying large language models (LLMs) on resource-constrained edge devices. However, fine-tuning quantized models presents significant challenges, primarily stemming from: First, the mismatch in data types between the low-precision quantized weights (e.g., 4-bit) and the high-precision adaptation weights (e.g., 16-bit). This mismatch limits the computational efficiency advantage offered by quantized weights during inference. Second, potential accuracy degradation when merging these high-precision adaptation weights into the low-precision quantized weights, as the adaptation weights often necessitate approximation or truncation. Third, as far as we know, no existing methods support the lossless merging of adaptation while adjusting all quantized weights. To address these challenges, we introduce lossless ternary adaptation for quantization-aware fine-tuning (LoTA-QAF). This is a novel fine-tuning method specifically designed for quantized LLMs, enabling the lossless merging of ternary adaptation weights into quantized weights and the adjustment of all quantized weights. LoTA-QAF operates through a combination of: i) A custom-designed ternary adaptation (TA) that aligns ternary weights with the quantization grid and uses these ternary weights to adjust quantized weights. ii) A TA-based mechanism that enables the lossless merging of adaptation weights. iii) Ternary signed gradient descent (t-SignSGD) for updating the TA weights. We apply LoTA-QAF to Llama-3.1/3.3 and Qwen-2.5 model families and validate its effectiveness on several downstream tasks. On the MMLU benchmark, our method effectively recovers performance for quantized models, surpassing 16-bit LoRA by up to 5.14\%. For task-specific fine-tuning, 16-bit LoRA achieves superior results, but LoTA-QAF still outperforms other methods.
Correlation Ratio for Unsupervised Learning of Multi-modal Deformable Registration
Apr 16, 2025Abstract:In recent years, unsupervised learning for deformable image registration has been a major research focus. This approach involves training a registration network using pairs of moving and fixed images, along with a loss function that combines an image similarity measure and deformation regularization. For multi-modal image registration tasks, the correlation ratio has been a widely-used image similarity measure historically, yet it has been underexplored in current deep learning methods. Here, we propose a differentiable correlation ratio to use as a loss function for learning-based multi-modal deformable image registration. This approach extends the traditionally non-differentiable implementation of the correlation ratio by using the Parzen windowing approximation, enabling backpropagation with deep neural networks. We validated the proposed correlation ratio on a multi-modal neuroimaging dataset. In addition, we established a Bayesian training framework to study how the trade-off between the deformation regularizer and similarity measures, including mutual information and our proposed correlation ratio, affects the registration performance.
SANA 1.5: Efficient Scaling of Training-Time and Inference-Time Compute in Linear Diffusion Transformer
Jan 30, 2025



Abstract:This paper presents SANA-1.5, a linear Diffusion Transformer for efficient scaling in text-to-image generation. Building upon SANA-1.0, we introduce three key innovations: (1) Efficient Training Scaling: A depth-growth paradigm that enables scaling from 1.6B to 4.8B parameters with significantly reduced computational resources, combined with a memory-efficient 8-bit optimizer. (2) Model Depth Pruning: A block importance analysis technique for efficient model compression to arbitrary sizes with minimal quality loss. (3) Inference-time Scaling: A repeated sampling strategy that trades computation for model capacity, enabling smaller models to match larger model quality at inference time. Through these strategies, SANA-1.5 achieves a text-image alignment score of 0.72 on GenEval, which can be further improved to 0.80 through inference scaling, establishing a new SoTA on GenEval benchmark. These innovations enable efficient model scaling across different compute budgets while maintaining high quality, making high-quality image generation more accessible.
MobileH2R: Learning Generalizable Human to Mobile Robot Handover Exclusively from Scalable and Diverse Synthetic Data
Jan 08, 2025



Abstract:This paper introduces MobileH2R, a framework for learning generalizable vision-based human-to-mobile-robot (H2MR) handover skills. Unlike traditional fixed-base handovers, this task requires a mobile robot to reliably receive objects in a large workspace enabled by its mobility. Our key insight is that generalizable handover skills can be developed in simulators using high-quality synthetic data, without the need for real-world demonstrations. To achieve this, we propose a scalable pipeline for generating diverse synthetic full-body human motion data, an automated method for creating safe and imitation-friendly demonstrations, and an efficient 4D imitation learning method for distilling large-scale demonstrations into closed-loop policies with base-arm coordination. Experimental evaluations in both simulators and the real world show significant improvements (at least +15% success rate) over baseline methods in all cases. Experiments also validate that large-scale and diverse synthetic data greatly enhances robot learning, highlighting our scalable framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge