Harrison Bai
A multitask framework for automated interpretation of multi-frame right upper quadrant ultrasound in clinical decision support
Jan 17, 2026Abstract:Ultrasound is a cornerstone of emergency and hepatobiliary imaging, yet its interpretation remains highly operator-dependent and time-sensitive. Here, we present a multitask vision-language agent (VLM) developed to assist with comprehensive right upper quadrant (RUQ) ultrasound interpretation across the full diagnostic workflow. The system was trained on a large, multi-center dataset comprising a primary cohort from Johns Hopkins Medical Institutions (9,189 cases, 594,099 images) and externally validated on cohorts from Stanford University (108 cases, 3,240 images) and a major Chinese medical center (257 cases, 3,178 images). Built on the Qwen2.5-VL-7B architecture, the agent integrates frame-level visual understanding with report-grounded language reasoning to perform three tasks: (i) classification of 18 hepatobiliary and gallbladder conditions, (ii) generation of clinically coherent diagnostic reports, and (iii) surgical decision support based on ultrasound findings and clinical data. The model achieved high diagnostic accuracy across all tasks, generated reports that were indistinguishable from expert-written versions in blinded evaluations, and demonstrated superior factual accuracy and information density on content-based metrics. The agent further identified patients requiring cholecystectomy with high precision, supporting real-time decision-making. These results highlight the potential of generalist vision-language models to improve diagnostic consistency, reporting efficiency, and surgical triage in real-world ultrasound practice.
Dataset and Benchmark for Enhancing Critical Retained Foreign Object Detection
Jul 09, 2025Abstract:Critical retained foreign objects (RFOs), including surgical instruments like sponges and needles, pose serious patient safety risks and carry significant financial and legal implications for healthcare institutions. Detecting critical RFOs using artificial intelligence remains challenging due to their rarity and the limited availability of chest X-ray datasets that specifically feature critical RFOs cases. Existing datasets only contain non-critical RFOs, like necklace or zipper, further limiting their utility for developing clinically impactful detection algorithms. To address these limitations, we introduce "Hopkins RFOs Bench", the first and largest dataset of its kind, containing 144 chest X-ray images of critical RFO cases collected over 18 years from the Johns Hopkins Health System. Using this dataset, we benchmark several state-of-the-art object detection models, highlighting the need for enhanced detection methodologies for critical RFO cases. Recognizing data scarcity challenges, we further explore image synthetic methods to bridge this gap. We evaluate two advanced synthetic image methods, DeepDRR-RFO, a physics-based method, and RoentGen-RFO, a diffusion-based method, for creating realistic radiographs featuring critical RFOs. Our comprehensive analysis identifies the strengths and limitations of each synthetic method, providing insights into effectively utilizing synthetic data to enhance model training. The Hopkins RFOs Bench and our findings significantly advance the development of reliable, generalizable AI-driven solutions for detecting critical RFOs in clinical chest X-rays.
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025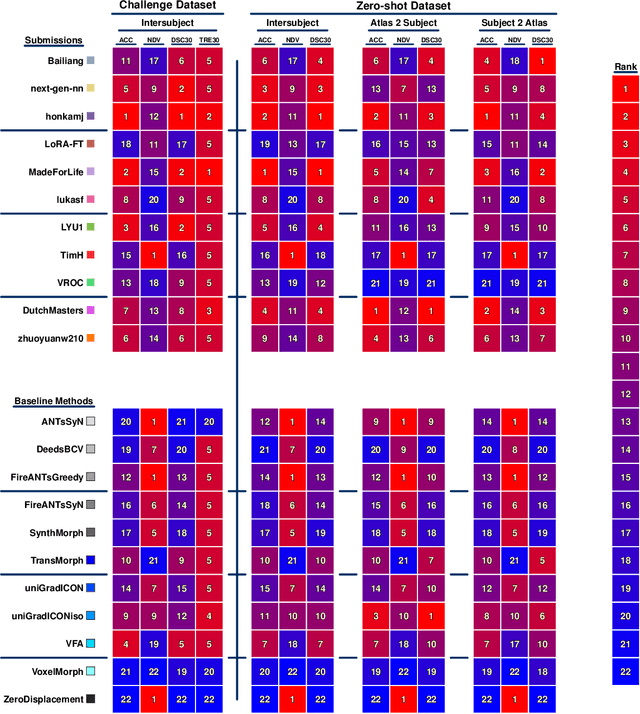
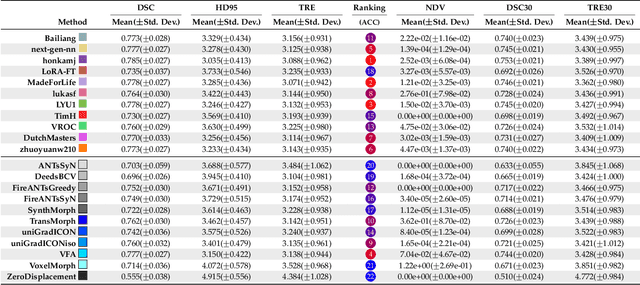
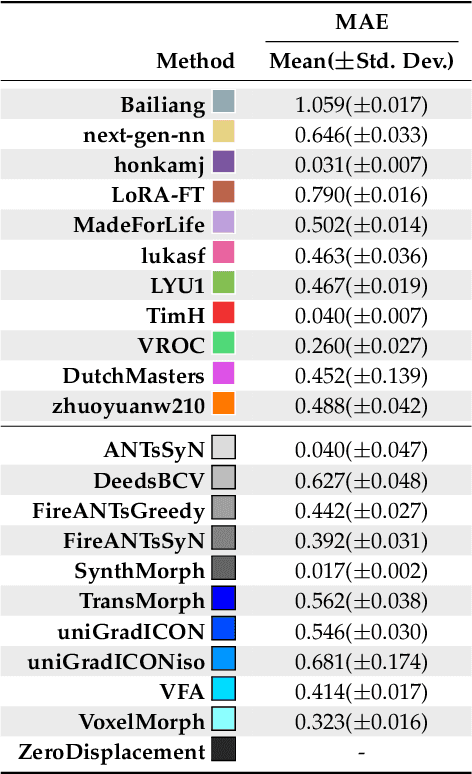
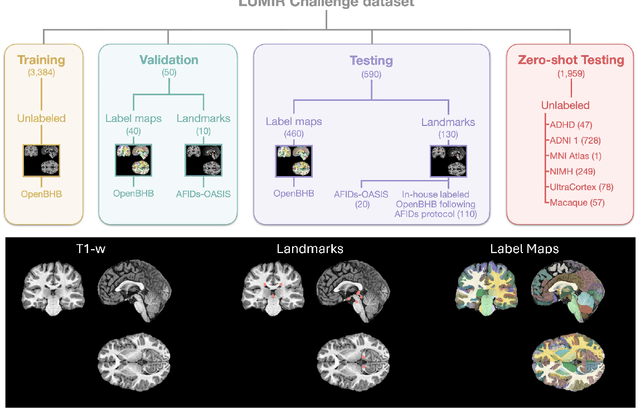
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
Unsupervised learning of spatially varying regularization for diffeomorphic image registration
Dec 23, 2024Abstract:Spatially varying regularization accommodates the deformation variations that may be necessary for different anatomical regions during deformable image registration. Historically, optimization-based registration models have harnessed spatially varying regularization to address anatomical subtleties. However, most modern deep learning-based models tend to gravitate towards spatially invariant regularization, wherein a homogenous regularization strength is applied across the entire image, potentially disregarding localized variations. In this paper, we propose a hierarchical probabilistic model that integrates a prior distribution on the deformation regularization strength, enabling the end-to-end learning of a spatially varying deformation regularizer directly from the data. The proposed method is straightforward to implement and easily integrates with various registration network architectures. Additionally, automatic tuning of hyperparameters is achieved through Bayesian optimization, allowing efficient identification of optimal hyperparameters for any given registration task. Comprehensive evaluations on publicly available datasets demonstrate that the proposed method significantly improves registration performance and enhances the interpretability of deep learning-based registration, all while maintaining smooth deformations.
Structural Entities Extraction and Patient Indications Incorporation for Chest X-ray Report Generation
May 23, 2024



Abstract:The automated generation of imaging reports proves invaluable in alleviating the workload of radiologists. A clinically applicable reports generation algorithm should demonstrate its effectiveness in producing reports that accurately describe radiology findings and attend to patient-specific indications. In this paper, we introduce a novel method, \textbf{S}tructural \textbf{E}ntities extraction and patient indications \textbf{I}ncorporation (SEI) for chest X-ray report generation. Specifically, we employ a structural entities extraction (SEE) approach to eliminate presentation-style vocabulary in reports and improve the quality of factual entity sequences. This reduces the noise in the following cross-modal alignment module by aligning X-ray images with factual entity sequences in reports, thereby enhancing the precision of cross-modal alignment and further aiding the model in gradient-free retrieval of similar historical cases. Subsequently, we propose a cross-modal fusion network to integrate information from X-ray images, similar historical cases, and patient-specific indications. This process allows the text decoder to attend to discriminative features of X-ray images, assimilate historical diagnostic information from similar cases, and understand the examination intention of patients. This, in turn, assists in triggering the text decoder to produce high-quality reports. Experiments conducted on MIMIC-CXR validate the superiority of SEI over state-of-the-art approaches on both natural language generation and clinical efficacy metrics.
Multi-modality Regional Alignment Network for Covid X-Ray Survival Prediction and Report Generation
May 23, 2024Abstract:In response to the worldwide COVID-19 pandemic, advanced automated technologies have emerged as valuable tools to aid healthcare professionals in managing an increased workload by improving radiology report generation and prognostic analysis. This study proposes Multi-modality Regional Alignment Network (MRANet), an explainable model for radiology report generation and survival prediction that focuses on high-risk regions. By learning spatial correlation in the detector, MRANet visually grounds region-specific descriptions, providing robust anatomical regions with a completion strategy. The visual features of each region are embedded using a novel survival attention mechanism, offering spatially and risk-aware features for sentence encoding while maintaining global coherence across tasks. A cross LLMs alignment is employed to enhance the image-to-text transfer process, resulting in sentences rich with clinical detail and improved explainability for radiologist. Multi-center experiments validate both MRANet's overall performance and each module's composition within the model, encouraging further advancements in radiology report generation research emphasizing clinical interpretation and trustworthiness in AI models applied to medical studies. The code is available at https://github.com/zzs95/MRANet.
Region-specific Risk Quantification for Interpretable Prognosis of COVID-19
May 05, 2024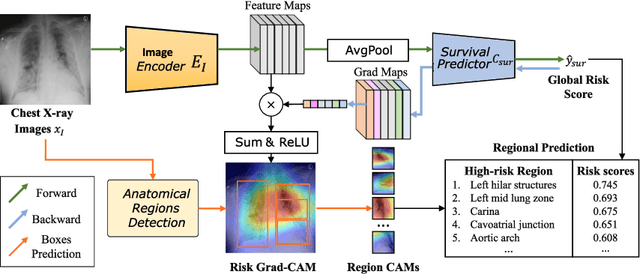
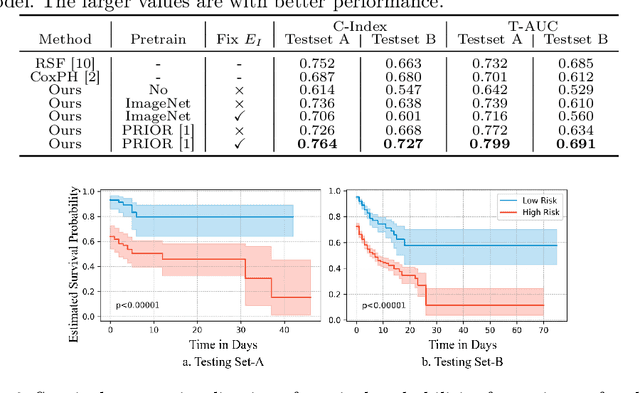
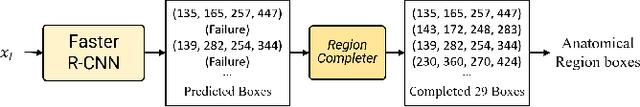
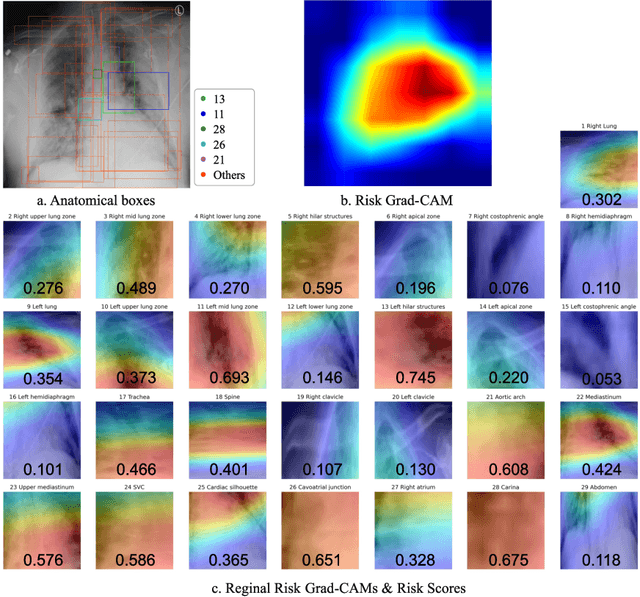
Abstract:The COVID-19 pandemic has strained global public health, necessitating accurate diagnosis and intervention to control disease spread and reduce mortality rates. This paper introduces an interpretable deep survival prediction model designed specifically for improved understanding and trust in COVID-19 prognosis using chest X-ray (CXR) images. By integrating a large-scale pretrained image encoder, Risk-specific Grad-CAM, and anatomical region detection techniques, our approach produces regional interpretable outcomes that effectively capture essential disease features while focusing on rare but critical abnormal regions. Our model's predictive results provide enhanced clarity and transparency through risk area localization, enabling clinicians to make informed decisions regarding COVID-19 diagnosis with better understanding of prognostic insights. We evaluate the proposed method on a multi-center survival dataset and demonstrate its effectiveness via quantitative and qualitative assessments, achieving superior C-indexes (0.764 and 0.727) and time-dependent AUCs (0.799 and 0.691). These results suggest that our explainable deep survival prediction model surpasses traditional survival analysis methods in risk prediction, improving interpretability for clinical decision making and enhancing AI system trustworthiness.
SMC-UDA: Structure-Modal Constraint for Unsupervised Cross-Domain Renal Segmentation
Jun 14, 2023Abstract:Medical image segmentation based on deep learning often fails when deployed on images from a different domain. The domain adaptation methods aim to solve domain-shift challenges, but still face some problems. The transfer learning methods require annotation on the target domain, and the generative unsupervised domain adaptation (UDA) models ignore domain-specific representations, whose generated quality highly restricts segmentation performance. In this study, we propose a novel Structure-Modal Constrained (SMC) UDA framework based on a discriminative paradigm and introduce edge structure as a bridge between domains. The proposed multi-modal learning backbone distills structure information from image texture to distinguish domain-invariant edge structure. With the structure-constrained self-learning and progressive ROI, our methods segment the kidney by locating the 3D spatial structure of the edge. We evaluated SMC-UDA on public renal segmentation datasets, adapting from the labeled source domain (CT) to the unlabeled target domain (CT/MRI). The experiments show that our proposed SMC-UDA has a strong generalization and outperforms generative UDA methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge