Zhicheng Jiao
A multitask framework for automated interpretation of multi-frame right upper quadrant ultrasound in clinical decision support
Jan 17, 2026Abstract:Ultrasound is a cornerstone of emergency and hepatobiliary imaging, yet its interpretation remains highly operator-dependent and time-sensitive. Here, we present a multitask vision-language agent (VLM) developed to assist with comprehensive right upper quadrant (RUQ) ultrasound interpretation across the full diagnostic workflow. The system was trained on a large, multi-center dataset comprising a primary cohort from Johns Hopkins Medical Institutions (9,189 cases, 594,099 images) and externally validated on cohorts from Stanford University (108 cases, 3,240 images) and a major Chinese medical center (257 cases, 3,178 images). Built on the Qwen2.5-VL-7B architecture, the agent integrates frame-level visual understanding with report-grounded language reasoning to perform three tasks: (i) classification of 18 hepatobiliary and gallbladder conditions, (ii) generation of clinically coherent diagnostic reports, and (iii) surgical decision support based on ultrasound findings and clinical data. The model achieved high diagnostic accuracy across all tasks, generated reports that were indistinguishable from expert-written versions in blinded evaluations, and demonstrated superior factual accuracy and information density on content-based metrics. The agent further identified patients requiring cholecystectomy with high precision, supporting real-time decision-making. These results highlight the potential of generalist vision-language models to improve diagnostic consistency, reporting efficiency, and surgical triage in real-world ultrasound practice.
CheXPO-v2: Preference Optimization for Chest X-ray VLMs with Knowledge Graph Consistency
Dec 19, 2025Abstract:Medical Vision-Language Models (VLMs) are prone to hallucinations, compromising clinical reliability. While reinforcement learning methods like Group Relative Policy Optimization (GRPO) offer a low-cost alignment solution, their reliance on sparse, outcome-based rewards inadvertently encourages models to "overthink" -- generating verbose, convoluted, and unverifiable Chain-of-Thought reasoning to justify answers. This focus on outcomes obscures factual errors and poses significant safety risks. To address this, we propose CheXPO-v2, a novel alignment framework that shifts from outcome to process supervision. Our core innovation is a Knowledge Graph Consistency Reward mechanism driven by Entity-Relation Matching. By explicitly parsing reasoning steps into structured "Disease, Relation, Anatomy" triplets, we provide fine-grained supervision that penalizes incoherent logic and hallucinations at the atomic level. Integrating this with a hard-example mining strategy, our approach significantly outperforms GRPO and state-of-the-art models on benchmarks like MIMIC-CXR-VQA. Crucially, CheXPO-v2 achieves new state-of-the-art accuracy using only 5k samples, demonstrating exceptional data efficiency while producing clinically sound and verifiable reasoning. The project source code is publicly available at: https://github.com/ecoxial2007/CheX-Phi4MM.
Abn-BLIP: Abnormality-aligned Bootstrapping Language-Image Pre-training for Pulmonary Embolism Diagnosis and Report Generation from CTPA
Mar 03, 2025Abstract:Medical imaging plays a pivotal role in modern healthcare, with computed tomography pulmonary angiography (CTPA) being a critical tool for diagnosing pulmonary embolism and other thoracic conditions. However, the complexity of interpreting CTPA scans and generating accurate radiology reports remains a significant challenge. This paper introduces Abn-BLIP (Abnormality-aligned Bootstrapping Language-Image Pretraining), an advanced diagnosis model designed to align abnormal findings to generate the accuracy and comprehensiveness of radiology reports. By leveraging learnable queries and cross-modal attention mechanisms, our model demonstrates superior performance in detecting abnormalities, reducing missed findings, and generating structured reports compared to existing methods. Our experiments show that Abn-BLIP outperforms state-of-the-art medical vision-language models and 3D report generation methods in both accuracy and clinical relevance. These results highlight the potential of integrating multimodal learning strategies for improving radiology reporting. The source code is available at https://github.com/zzs95/abn-blip.
Enhanced Contrastive Learning with Multi-view Longitudinal Data for Chest X-ray Report Generation
Feb 27, 2025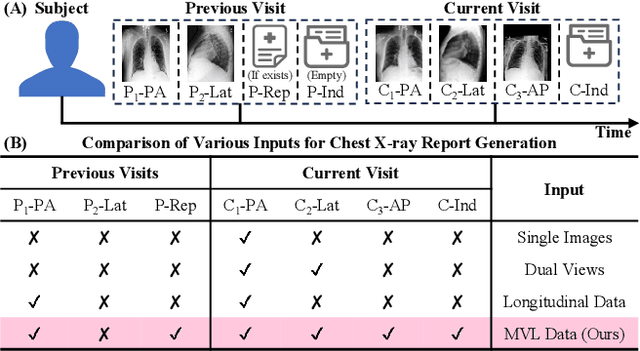
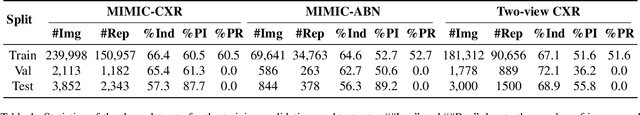
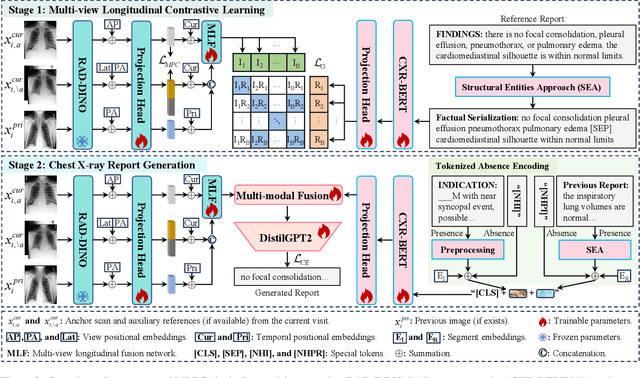
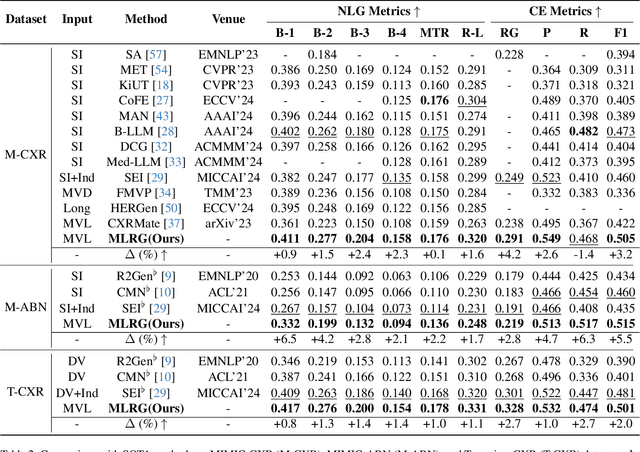
Abstract:Automated radiology report generation offers an effective solution to alleviate radiologists' workload. However, most existing methods focus primarily on single or fixed-view images to model current disease conditions, which limits diagnostic accuracy and overlooks disease progression. Although some approaches utilize longitudinal data to track disease progression, they still rely on single images to analyze current visits. To address these issues, we propose enhanced contrastive learning with Multi-view Longitudinal data to facilitate chest X-ray Report Generation, named MLRG. Specifically, we introduce a multi-view longitudinal contrastive learning method that integrates spatial information from current multi-view images and temporal information from longitudinal data. This method also utilizes the inherent spatiotemporal information of radiology reports to supervise the pre-training of visual and textual representations. Subsequently, we present a tokenized absence encoding technique to flexibly handle missing patient-specific prior knowledge, allowing the model to produce more accurate radiology reports based on available prior knowledge. Extensive experiments on MIMIC-CXR, MIMIC-ABN, and Two-view CXR datasets demonstrate that our MLRG outperforms recent state-of-the-art methods, achieving a 2.3% BLEU-4 improvement on MIMIC-CXR, a 5.5% F1 score improvement on MIMIC-ABN, and a 2.7% F1 RadGraph improvement on Two-view CXR.
Optimizing Prompt Strategies for SAM: Advancing lesion Segmentation Across Diverse Medical Imaging Modalities
Dec 28, 2024



Abstract:Purpose: To evaluate various Segmental Anything Model (SAM) prompt strategies across four lesions datasets and to subsequently develop a reinforcement learning (RL) agent to optimize SAM prompt placement. Materials and Methods: This retrospective study included patients with four independent ovarian, lung, renal, and breast tumor datasets. Manual segmentation and SAM-assisted segmentation were performed for all lesions. A RL model was developed to predict and select SAM points to maximize segmentation performance. Statistical analysis of segmentation was conducted using pairwise t-tests. Results: Results show that increasing the number of prompt points significantly improves segmentation accuracy, with Dice coefficients rising from 0.272 for a single point to 0.806 for five or more points in ovarian tumors. The prompt location also influenced performance, with surface and union-based prompts outperforming center-based prompts, achieving mean Dice coefficients of 0.604 and 0.724 for ovarian and breast tumors, respectively. The RL agent achieved a peak Dice coefficient of 0.595 for ovarian tumors, outperforming random and alternative RL strategies. Additionally, it significantly reduced segmentation time, achieving a nearly 10-fold improvement compared to manual methods using SAM. Conclusion: While increased SAM prompts and non-centered prompts generally improved segmentation accuracy, each pathology and modality has specific optimal thresholds and placement strategies. Our RL agent achieved superior performance compared to other agents while achieving a significant reduction in segmentation time.
Optimizing prompt strategies for the Segment Anything Model are explored, focusing on prompt location, number, and reinforcement learning-based agent for prompt placement across four lesion datasets
Dec 23, 2024



Abstract:Purpose: To evaluate various Segmental Anything Model (SAM) prompt strategies across four lesions datasets and to subsequently develop a reinforcement learning (RL) agent to optimize SAM prompt placement. Materials and Methods: This retrospective study included patients with four independent ovarian, lung, renal, and breast tumor datasets. Manual segmentation and SAM-assisted segmentation were performed for all lesions. A RL model was developed to predict and select SAM points to maximize segmentation performance. Statistical analysis of segmentation was conducted using pairwise t-tests. Results: Results show that increasing the number of prompt points significantly improves segmentation accuracy, with Dice coefficients rising from 0.272 for a single point to 0.806 for five or more points in ovarian tumors. The prompt location also influenced performance, with surface and union-based prompts outperforming center-based prompts, achieving mean Dice coefficients of 0.604 and 0.724 for ovarian and breast tumors, respectively. The RL agent achieved a peak Dice coefficient of 0.595 for ovarian tumors, outperforming random and alternative RL strategies. Additionally, it significantly reduced segmentation time, achieving a nearly 10-fold improvement compared to manual methods using SAM. Conclusion: While increased SAM prompts and non-centered prompts generally improved segmentation accuracy, each pathology and modality has specific optimal thresholds and placement strategies. Our RL agent achieved superior performance compared to other agents while achieving a significant reduction in segmentation time.
MCL: Multi-view Enhanced Contrastive Learning for Chest X-ray Report Generation
Nov 15, 2024



Abstract:Radiology reports are crucial for planning treatment strategies and enhancing doctor-patient communication, yet manually writing these reports is burdensome for radiologists. While automatic report generation offers a solution, existing methods often rely on single-view radiographs, limiting diagnostic accuracy. To address this problem, we propose MCL, a Multi-view enhanced Contrastive Learning method for chest X-ray report generation. Specifically, we first introduce multi-view enhanced contrastive learning for visual representation by maximizing agreements between multi-view radiographs and their corresponding report. Subsequently, to fully exploit patient-specific indications (e.g., patient's symptoms) for report generation, we add a transitional ``bridge" for missing indications to reduce embedding space discrepancies caused by their presence or absence. Additionally, we construct Multi-view CXR and Two-view CXR datasets from public sources to support research on multi-view report generation. Our proposed MCL surpasses recent state-of-the-art methods across multiple datasets, achieving a 5.0% F1 RadGraph improvement on MIMIC-CXR, a 7.3% BLEU-1 improvement on MIMIC-ABN, a 3.1% BLEU-4 improvement on Multi-view CXR, and an 8.2% F1 CheXbert improvement on Two-view CXR.
Pulmonary Embolism Mortality Prediction Using Multimodal Learning Based on Computed Tomography Angiography and Clinical Data
Jun 03, 2024Abstract:Purpose: Pulmonary embolism (PE) is a significant cause of mortality in the United States. The objective of this study is to implement deep learning (DL) models using Computed Tomography Pulmonary Angiography (CTPA), clinical data, and PE Severity Index (PESI) scores to predict PE mortality. Materials and Methods: 918 patients (median age 64 years, range 13-99 years, 52% female) with 3,978 CTPAs were identified via retrospective review across three institutions. To predict survival, an AI model was used to extract disease-related imaging features from CTPAs. Imaging features and/or clinical variables were then incorporated into DL models to predict survival outcomes. Four models were developed as follows: (1) using CTPA imaging features only; (2) using clinical variables only; (3) multimodal, integrating both CTPA and clinical variables; and (4) multimodal fused with calculated PESI score. Performance and contribution from each modality were evaluated using concordance index (c-index) and Net Reclassification Improvement, respectively. Performance was compared to PESI predictions using the Wilcoxon signed-rank test. Kaplan-Meier analysis was performed to stratify patients into high- and low-risk groups. Additional factor-risk analysis was conducted to account for right ventricular (RV) dysfunction. Results: For both data sets, the PESI-fused and multimodal models achieved higher c-indices than PESI alone. Following stratification of patients into high- and low-risk groups by multimodal and PESI-fused models, mortality outcomes differed significantly (both p<0.001). A strong correlation was found between high-risk grouping and RV dysfunction. Conclusions: Multiomic DL models incorporating CTPA features, clinical data, and PESI achieved higher c-indices than PESI alone for PE survival prediction.
MindSemantix: Deciphering Brain Visual Experiences with a Brain-Language Model
May 29, 2024Abstract:Deciphering the human visual experience through brain activities captured by fMRI represents a compelling and cutting-edge challenge in the field of neuroscience research. Compared to merely predicting the viewed image itself, decoding brain activity into meaningful captions provides a higher-level interpretation and summarization of visual information, which naturally enhances the application flexibility in real-world situations. In this work, we introduce MindSemantix, a novel multi-modal framework that enables LLMs to comprehend visually-evoked semantic content in brain activity. Our MindSemantix explores a more ideal brain captioning paradigm by weaving LLMs into brain activity analysis, crafting a seamless, end-to-end Brain-Language Model. To effectively capture semantic information from brain responses, we propose Brain-Text Transformer, utilizing a Brain Q-Former as its core architecture. It integrates a pre-trained brain encoder with a frozen LLM to achieve multi-modal alignment of brain-vision-language and establish a robust brain-language correspondence. To enhance the generalizability of neural representations, we pre-train our brain encoder on a large-scale, cross-subject fMRI dataset using self-supervised learning techniques. MindSemantix provides more feasibility to downstream brain decoding tasks such as stimulus reconstruction. Conditioned by MindSemantix captioning, our framework facilitates this process by integrating with advanced generative models like Stable Diffusion and excels in understanding brain visual perception. MindSemantix generates high-quality captions that are deeply rooted in the visual and semantic information derived from brain activity. This approach has demonstrated substantial quantitative improvements over prior art. Our code will be released.
Structural Entities Extraction and Patient Indications Incorporation for Chest X-ray Report Generation
May 23, 2024



Abstract:The automated generation of imaging reports proves invaluable in alleviating the workload of radiologists. A clinically applicable reports generation algorithm should demonstrate its effectiveness in producing reports that accurately describe radiology findings and attend to patient-specific indications. In this paper, we introduce a novel method, \textbf{S}tructural \textbf{E}ntities extraction and patient indications \textbf{I}ncorporation (SEI) for chest X-ray report generation. Specifically, we employ a structural entities extraction (SEE) approach to eliminate presentation-style vocabulary in reports and improve the quality of factual entity sequences. This reduces the noise in the following cross-modal alignment module by aligning X-ray images with factual entity sequences in reports, thereby enhancing the precision of cross-modal alignment and further aiding the model in gradient-free retrieval of similar historical cases. Subsequently, we propose a cross-modal fusion network to integrate information from X-ray images, similar historical cases, and patient-specific indications. This process allows the text decoder to attend to discriminative features of X-ray images, assimilate historical diagnostic information from similar cases, and understand the examination intention of patients. This, in turn, assists in triggering the text decoder to produce high-quality reports. Experiments conducted on MIMIC-CXR validate the superiority of SEI over state-of-the-art approaches on both natural language generation and clinical efficacy metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge