Qingyun Wang
oMeBench: Towards Robust Benchmarking of LLMs in Organic Mechanism Elucidation and Reasoning
Oct 09, 2025Abstract:Organic reaction mechanisms are the stepwise elementary reactions by which reactants form intermediates and products, and are fundamental to understanding chemical reactivity and designing new molecules and reactions. Although large language models (LLMs) have shown promise in understanding chemical tasks such as synthesis design, it is unclear to what extent this reflects genuine chemical reasoning capabilities, i.e., the ability to generate valid intermediates, maintain chemical consistency, and follow logically coherent multi-step pathways. We address this by introducing oMeBench, the first large-scale, expert-curated benchmark for organic mechanism reasoning in organic chemistry. It comprises over 10,000 annotated mechanistic steps with intermediates, type labels, and difficulty ratings. Furthermore, to evaluate LLM capability more precisely and enable fine-grained scoring, we propose oMeS, a dynamic evaluation framework that combines step-level logic and chemical similarity. We analyze the performance of state-of-the-art LLMs, and our results show that although current models display promising chemical intuition, they struggle with correct and consistent multi-step reasoning. Notably, we find that using prompting strategy and fine-tuning a specialist model on our proposed dataset increases performance by 50% over the leading closed-source model. We hope that oMeBench will serve as a rigorous foundation for advancing AI systems toward genuine chemical reasoning.
Affine Modulation-based Audiogram Fusion Network for Joint Noise Reduction and Hearing Loss Compensation
Sep 09, 2025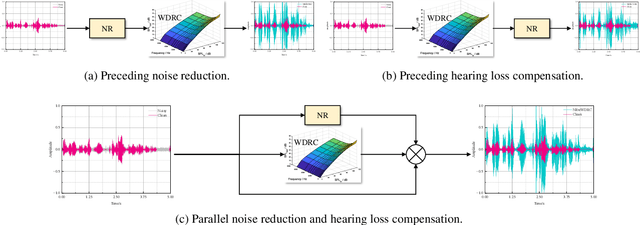
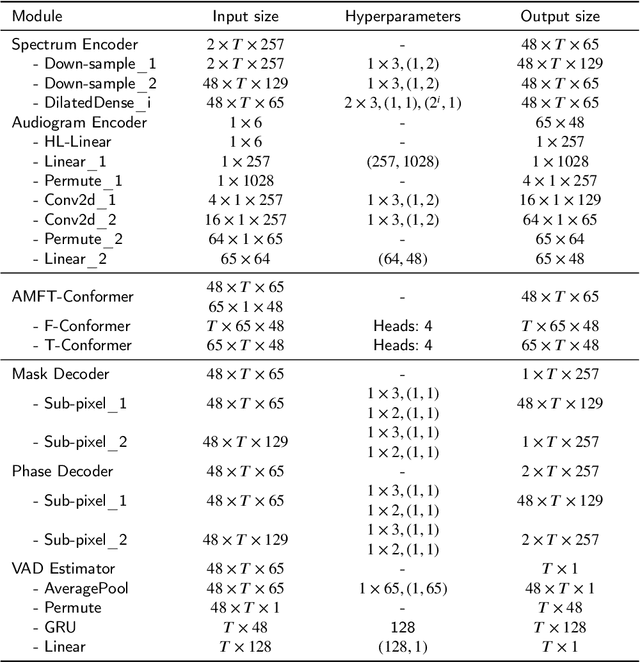
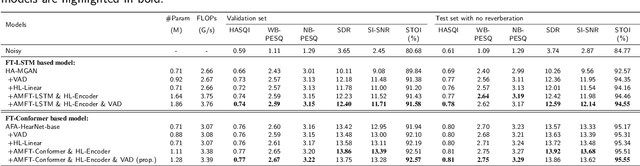
Abstract:Hearing aids (HAs) are widely used to provide personalized speech enhancement (PSE) services, improving the quality of life for individuals with hearing loss. However, HA performance significantly declines in noisy environments as it treats noise reduction (NR) and hearing loss compensation (HLC) as separate tasks. This separation leads to a lack of systematic optimization, overlooking the interactions between these two critical tasks, and increases the system complexity. To address these challenges, we propose a novel audiogram fusion network, named AFN-HearNet, which simultaneously tackles the NR and HLC tasks by fusing cross-domain audiogram and spectrum features. We propose an audiogram-specific encoder that transforms the sparse audiogram profile into a deep representation, addressing the alignment problem of cross-domain features prior to fusion. To incorporate the interactions between NR and HLC tasks, we propose the affine modulation-based audiogram fusion frequency-temporal Conformer that adaptively fuses these two features into a unified deep representation for speech reconstruction. Furthermore, we introduce a voice activity detection auxiliary training task to embed speech and non-speech patterns into the unified deep representation implicitly. We conduct comprehensive experiments across multiple datasets to validate the effectiveness of each proposed module. The results indicate that the AFN-HearNet significantly outperforms state-of-the-art in-context fusion joint models regarding key metrics such as HASQI and PESQ, achieving a considerable trade-off between performance and efficiency. The source code and data will be released at https://github.com/deepnetni/AFN-HearNet.
Atomic Reasoning for Scientific Table Claim Verification
Jun 08, 2025Abstract:Scientific texts often convey authority due to their technical language and complex data. However, this complexity can sometimes lead to the spread of misinformation. Non-experts are particularly susceptible to misleading claims based on scientific tables due to their high information density and perceived credibility. Existing table claim verification models, including state-of-the-art large language models (LLMs), often struggle with precise fine-grained reasoning, resulting in errors and a lack of precision in verifying scientific claims. Inspired by Cognitive Load Theory, we propose that enhancing a model's ability to interpret table-based claims involves reducing cognitive load by developing modular, reusable reasoning components (i.e., atomic skills). We introduce a skill-chaining schema that dynamically composes these skills to facilitate more accurate and generalizable reasoning with a reduced cognitive load. To evaluate this, we create SciAtomicBench, a cross-domain benchmark with fine-grained reasoning annotations. With only 350 fine-tuning examples, our model trained by atomic reasoning outperforms GPT-4o's chain-of-thought method, achieving state-of-the-art results with far less training data.
Explain Less, Understand More: Jargon Detection via Personalized Parameter-Efficient Fine-tuning
May 22, 2025
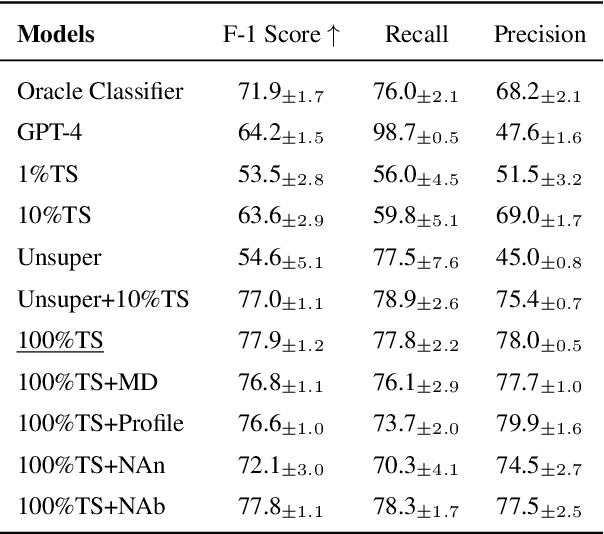
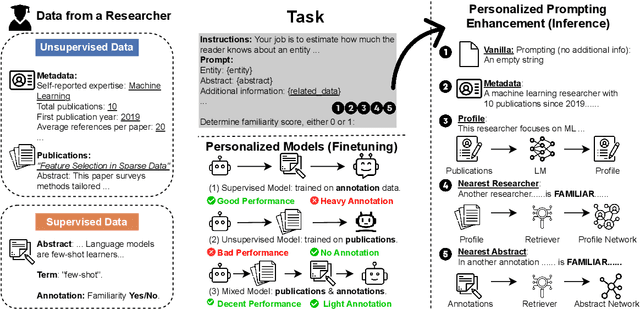

Abstract:Personalizing jargon detection and explanation is essential for making technical documents accessible to readers with diverse disciplinary backgrounds. However, tailoring models to individual users typically requires substantial annotation efforts and computational resources due to user-specific finetuning. To address this, we present a systematic study of personalized jargon detection, focusing on methods that are both efficient and scalable for real-world deployment. We explore two personalization strategies: (1) lightweight fine-tuning using Low-Rank Adaptation (LoRA) on open-source models, and (2) personalized prompting, which tailors model behavior at inference time without retaining. To reflect realistic constraints, we also investigate hybrid approaches that combine limited annotated data with unsupervised user background signals. Our personalized LoRA model outperforms GPT-4 by 21.4% in F1 score and exceeds the best performing oracle baseline by 8.3%. Remarkably, our method achieves comparable performance using only 10% of the annotated training data, demonstrating its practicality for resource-constrained settings. Our study offers the first work to systematically explore efficient, low-resource personalization of jargon detection using open-source language models, offering a practical path toward scalable, user-adaptive NLP system.
MAC-Tuning: LLM Multi-Compositional Problem Reasoning with Enhanced Knowledge Boundary Awareness
Apr 30, 2025Abstract:With the widespread application of large language models (LLMs), the issue of generating non-existing facts, known as hallucination, has garnered increasing attention. Previous research in enhancing LLM confidence estimation mainly focuses on the single problem setting. However, LLM awareness of its internal parameterized knowledge boundary under the more challenging multi-problem setting, which requires answering multiple problems accurately simultaneously, remains underexplored. To bridge this gap, we introduce a novel method, Multiple Answers and Confidence Stepwise Tuning (MAC-Tuning), that separates the learning of answer prediction and confidence estimation during fine-tuning on instruction data. Extensive experiments demonstrate that our method outperforms baselines by up to 25% in average precision.
CALM: Unleashing the Cross-Lingual Self-Aligning Ability of Language Model Question Answering
Jan 30, 2025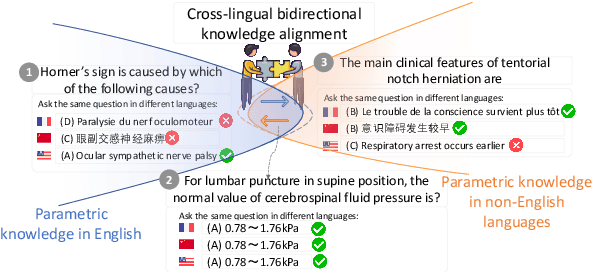



Abstract:Large Language Models (LLMs) are pretrained on extensive multilingual corpora to acquire both language-specific cultural knowledge and general knowledge. Ideally, while LLMs should provide consistent responses to culture-independent questions across languages, we observe significant performance disparities. To address this, we explore the Cross-Lingual Self-Aligning ability of Language Models (CALM) to align knowledge across languages. Specifically, for a given question, we sample multiple responses across different languages, and select the most self-consistent response as the target, leaving the remaining responses as negative examples. We then employ direct preference optimization (DPO) to align the model's knowledge across different languages. Evaluations on the MEDQA and X-CSQA datasets demonstrate CALM's effectiveness in enhancing cross-lingual knowledge question answering, both in zero-shot and retrieval augmented settings. We also found that increasing the number of languages involved in CALM training leads to even higher accuracy and consistency. We offer a qualitative analysis of how cross-lingual consistency can enhance knowledge alignment and explore the method's generalizability. The source code and data of this paper are available on GitHub.
Schema-Guided Culture-Aware Complex Event Simulation with Multi-Agent Role-Play
Oct 24, 2024



Abstract:Complex news events, such as natural disasters and socio-political conflicts, require swift responses from the government and society. Relying on historical events to project the future is insufficient as such events are sparse and do not cover all possible conditions and nuanced situations. Simulation of these complex events can help better prepare and reduce the negative impact. We develop a controllable complex news event simulator guided by both the event schema representing domain knowledge about the scenario and user-provided assumptions representing case-specific conditions. As event dynamics depend on the fine-grained social and cultural context, we further introduce a geo-diverse commonsense and cultural norm-aware knowledge enhancement component. To enhance the coherence of the simulation, apart from the global timeline of events, we take an agent-based approach to simulate the individual character states, plans, and actions. By incorporating the schema and cultural norms, our generated simulations achieve much higher coherence and appropriateness and are received favorably by participants from a humanitarian assistance organization.
Gene-Metabolite Association Prediction with Interactive Knowledge Transfer Enhanced Graph for Metabolite Production
Oct 24, 2024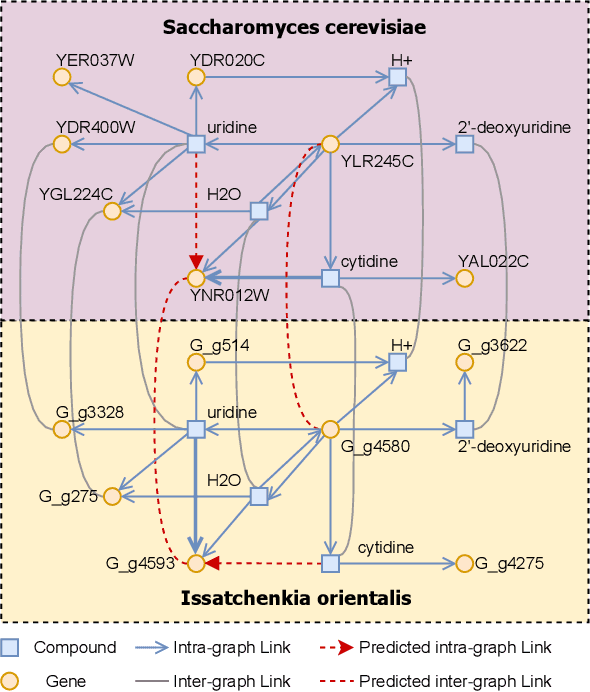
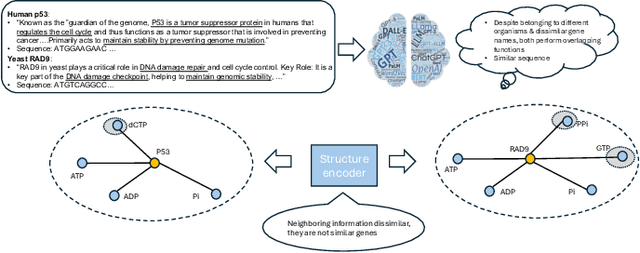
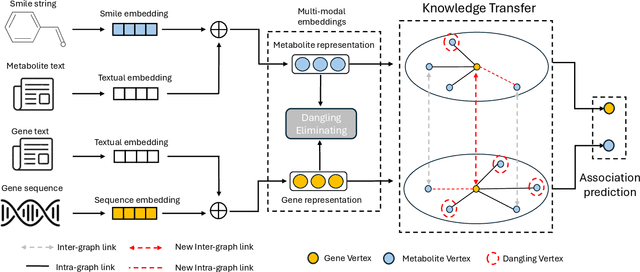
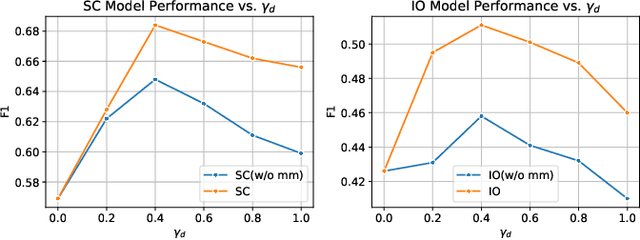
Abstract:In the rapidly evolving field of metabolic engineering, the quest for efficient and precise gene target identification for metabolite production enhancement presents significant challenges. Traditional approaches, whether knowledge-based or model-based, are notably time-consuming and labor-intensive, due to the vast scale of research literature and the approximation nature of genome-scale metabolic model (GEM) simulations. Therefore, we propose a new task, Gene-Metabolite Association Prediction based on metabolic graphs, to automate the process of candidate gene discovery for a given pair of metabolite and candidate-associated genes, as well as presenting the first benchmark containing 2474 metabolites and 1947 genes of two commonly used microorganisms Saccharomyces cerevisiae (SC) and Issatchenkia orientalis (IO). This task is challenging due to the incompleteness of the metabolic graphs and the heterogeneity among distinct metabolisms. To overcome these limitations, we propose an Interactive Knowledge Transfer mechanism based on Metabolism Graph (IKT4Meta), which improves the association prediction accuracy by integrating the knowledge from different metabolism graphs. First, to build a bridge between two graphs for knowledge transfer, we utilize Pretrained Language Models (PLMs) with external knowledge of genes and metabolites to help generate inter-graph links, significantly alleviating the impact of heterogeneity. Second, we propagate intra-graph links from different metabolic graphs using inter-graph links as anchors. Finally, we conduct the gene-metabolite association prediction based on the enriched metabolism graphs, which integrate the knowledge from multiple microorganisms. Experiments on both types of organisms demonstrate that our proposed methodology outperforms baselines by up to 12.3% across various link prediction frameworks.
MentalArena: Self-play Training of Language Models for Diagnosis and Treatment of Mental Health Disorders
Oct 09, 2024



Abstract:Mental health disorders are one of the most serious diseases in the world. Most people with such a disease lack access to adequate care, which highlights the importance of training models for the diagnosis and treatment of mental health disorders. However, in the mental health domain, privacy concerns limit the accessibility of personalized treatment data, making it challenging to build powerful models. In this paper, we introduce MentalArena, a self-play framework to train language models by generating domain-specific personalized data, where we obtain a better model capable of making a personalized diagnosis and treatment (as a therapist) and providing information (as a patient). To accurately model human-like mental health patients, we devise Symptom Encoder, which simulates a real patient from both cognition and behavior perspectives. To address intent bias during patient-therapist interactions, we propose Symptom Decoder to compare diagnosed symptoms with encoded symptoms, and dynamically manage the dialogue between patient and therapist according to the identified deviations. We evaluated MentalArena against 6 benchmarks, including biomedicalQA and mental health tasks, compared to 6 advanced models. Our models, fine-tuned on both GPT-3.5 and Llama-3-8b, significantly outperform their counterparts, including GPT-4o. We hope that our work can inspire future research on personalized care. Code is available in https://github.com/Scarelette/MentalArena/tree/main
Self-Correction is More than Refinement: A Learning Framework for Visual and Language Reasoning Tasks
Oct 05, 2024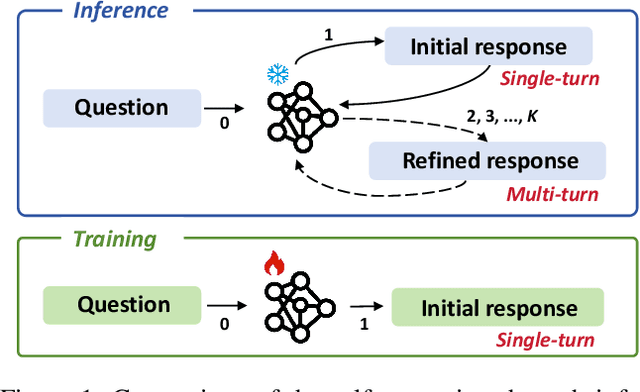
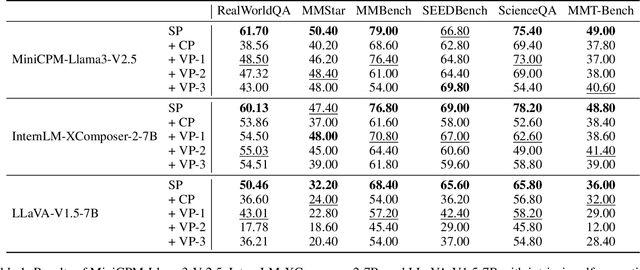
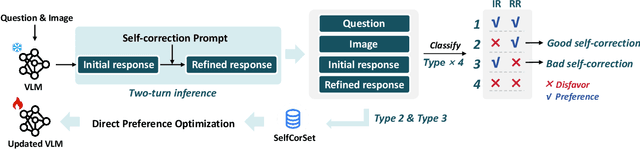
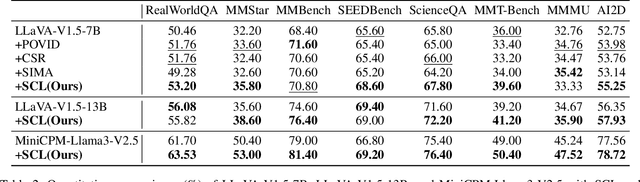
Abstract:While Vision-Language Models (VLMs) have shown remarkable abilities in visual and language reasoning tasks, they invariably generate flawed responses. Self-correction that instructs models to refine their outputs presents a promising solution to this issue. Previous studies have mainly concentrated on Large Language Models (LLMs), while the self-correction abilities of VLMs, particularly concerning both visual and linguistic information, remain largely unexamined. This study investigates the self-correction capabilities of VLMs during both inference and fine-tuning stages. We introduce a Self-Correction Learning (SCL) approach that enables VLMs to learn from their self-generated self-correction data through Direct Preference Optimization (DPO) without relying on external feedback, facilitating self-improvement. Specifically, we collect preferred and disfavored samples based on the correctness of initial and refined responses, which are obtained by two-turn self-correction with VLMs during the inference stage. Experimental results demonstrate that although VLMs struggle to self-correct effectively during iterative inference without additional fine-tuning and external feedback, they can enhance their performance and avoid previous mistakes through preference fine-tuning when their self-generated self-correction data are categorized into preferred and disfavored samples. This study emphasizes that self-correction is not merely a refinement process; rather, it should enhance the reasoning abilities of models through additional training, enabling them to generate high-quality responses directly without further refinement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge