Haoyu Dong
FORESTLLM: Large Language Models Make Random Forest Great on Few-shot Tabular Learning
Jan 16, 2026Abstract:Tabular data high-stakes critical decision-making in domains such as finance, healthcare, and scientific discovery. Yet, learning effectively from tabular data in few-shot settings, where labeled examples are scarce, remains a fundamental challenge. Traditional tree-based methods often falter in these regimes due to their reliance on statistical purity metrics, which become unstable and prone to overfitting with limited supervision. At the same time, direct applications of large language models (LLMs) often overlook its inherent structure, leading to suboptimal performance. To overcome these limitations, we propose FORESTLLM, a novel framework that unifies the structural inductive biases of decision forests with the semantic reasoning capabilities of LLMs. Crucially, FORESTLLM leverages the LLM only during training, treating it as an offline model designer that encodes rich, contextual knowledge into a lightweight, interpretable forest model, eliminating the need for LLM inference at test time. Our method is two-fold. First, we introduce a semantic splitting criterion in which the LLM evaluates candidate partitions based on their coherence over both labeled and unlabeled data, enabling the induction of more robust and generalizable tree structures under few-shot supervision. Second, we propose a one-time in-context inference mechanism for leaf node stabilization, where the LLM distills the decision path and its supporting examples into a concise, deterministic prediction, replacing noisy empirical estimates with semantically informed outputs. Across a diverse suite of few-shot classification and regression benchmarks, FORESTLLM achieves state-of-the-art performance.
LitVISTA: A Benchmark for Narrative Orchestration in Literary Text
Jan 10, 2026Abstract:Computational narrative analysis aims to capture rhythm, tension, and emotional dynamics in literary texts. Existing large language models can generate long stories but overly focus on causal coherence, neglecting the complex story arcs and orchestration inherent in human narratives. This creates a structural misalignment between model- and human-generated narratives. We propose VISTA Space, a high-dimensional representational framework for narrative orchestration that unifies human and model narrative perspectives. We further introduce LitVISTA, a structurally annotated benchmark grounded in literary texts, enabling systematic evaluation of models' narrative orchestration capabilities. We conduct oracle evaluations on a diverse selection of frontier LLMs, including GPT, Claude, Grok, and Gemini. Results reveal systematic deficiencies: existing models fail to construct a unified global narrative view, struggling to jointly capture narrative function and structure. Furthermore, even advanced thinking modes yield only limited gains for such literary narrative understanding.
Closing the Reality Gap: Zero-Shot Sim-to-Real Deployment for Dexterous Force-Based Grasping and Manipulation
Jan 06, 2026Abstract:Human-like dexterous hands with multiple fingers offer human-level manipulation capabilities, but training control policies that can directly deploy on real hardware remains difficult due to contact-rich physics and imperfect actuation. We close this gap with a practical sim-to-real reinforcement learning (RL) framework that utilizes dense tactile feedback combined with joint torque sensing to explicitly regulate physical interactions. To enable effective sim-to-real transfer, we introduce (i) a computationally fast tactile simulation that computes distances between dense virtual tactile units and the object via parallel forward kinematics, providing high-rate, high-resolution touch signals needed by RL; (ii) a current-to-torque calibration that eliminates the need for torque sensors on dexterous hands by mapping motor current to joint torque; and (iii) actuator dynamics modeling to bridge the actuation gaps with randomization of non-ideal effects such as backlash, torque-speed saturation. Using an asymmetric actor-critic PPO pipeline trained entirely in simulation, our policies deploy directly to a five-finger hand. The resulting policies demonstrated two essential skills: (1) command-based, controllable grasp force tracking, and (2) reorientation of objects in the hand, both of which were robustly executed without fine-tuning on the robot. By combining tactile and torque in the observation space with effective sensing/actuation modeling, our system provides a practical solution to achieve reliable dexterous manipulation. To our knowledge, this is the first demonstration of controllable grasping on a multi-finger dexterous hand trained entirely in simulation and transferred zero-shot on real hardware.
Finch: Benchmarking Finance & Accounting across Spreadsheet-Centric Enterprise Workflows
Dec 19, 2025Abstract:We introduce a finance & accounting benchmark (Finch) for evaluating AI agents on real-world, enterprise-grade professional workflows -- interleaving data entry, structuring, formatting, web search, cross-file retrieval, calculation, modeling, validation, translation, visualization, and reporting. Finch is sourced from authentic enterprise workspaces at Enron (15,000 spreadsheets and 500,000 emails from 150 employees) and other financial institutions, preserving in-the-wild messiness across multimodal artifacts (text, tables, formulas, charts, code, and images) and spanning diverse domains such as budgeting, trading, and asset management. We propose a workflow construction process that combines LLM-assisted discovery with expert annotation: (1) LLM-assisted, expert-verified derivation of workflows from real-world email threads and version histories of spreadsheet files, and (2) meticulous expert annotation for workflows, requiring over 700 hours of domain-expert effort. This yields 172 composite workflows with 384 tasks, involving 1,710 spreadsheets with 27 million cells, along with PDFs and other artifacts, capturing the intrinsically messy, long-horizon, knowledge-intensive, and collaborative nature of real-world enterprise work. We conduct both human and automated evaluations of frontier AI systems including GPT 5.1, Claude Sonnet 4.5, Gemini 3 Pro, Grok 4, and Qwen 3 Max, and GPT 5.1 Pro spends 48 hours in total yet passes only 38.4% of workflows, while Claude Sonnet 4.5 passes just 25.0%. Comprehensive case studies further surface the challenges that real-world enterprise workflows pose for AI agents.
SheetBrain: A Neuro-Symbolic Agent for Accurate Reasoning over Complex and Large Spreadsheets
Oct 22, 2025Abstract:Understanding and reasoning over complex spreadsheets remain fundamental challenges for large language models (LLMs), which often struggle with accurately capturing the complex structure of tables and ensuring reasoning correctness. In this work, we propose SheetBrain, a neuro-symbolic dual workflow agent framework designed for accurate reasoning over tabular data, supporting both spreadsheet question answering and manipulation tasks. SheetBrain comprises three core modules: an understanding module, which produces a comprehensive overview of the spreadsheet - including sheet summary and query-based problem insight to guide reasoning; an execution module, which integrates a Python sandbox with preloaded table-processing libraries and an Excel helper toolkit for effective multi-turn reasoning; and a validation module, which verifies the correctness of reasoning and answers, triggering re-execution when necessary. We evaluate SheetBrain on multiple public tabular QA and manipulation benchmarks, and introduce SheetBench, a new benchmark targeting large, multi-table, and structurally complex spreadsheets. Experimental results show that SheetBrain significantly improves accuracy on both existing benchmarks and the more challenging scenarios presented in SheetBench. Our code is publicly available at https://github.com/microsoft/SheetBrain.
Jupiter: Enhancing LLM Data Analysis Capabilities via Notebook and Inference-Time Value-Guided Search
Sep 11, 2025

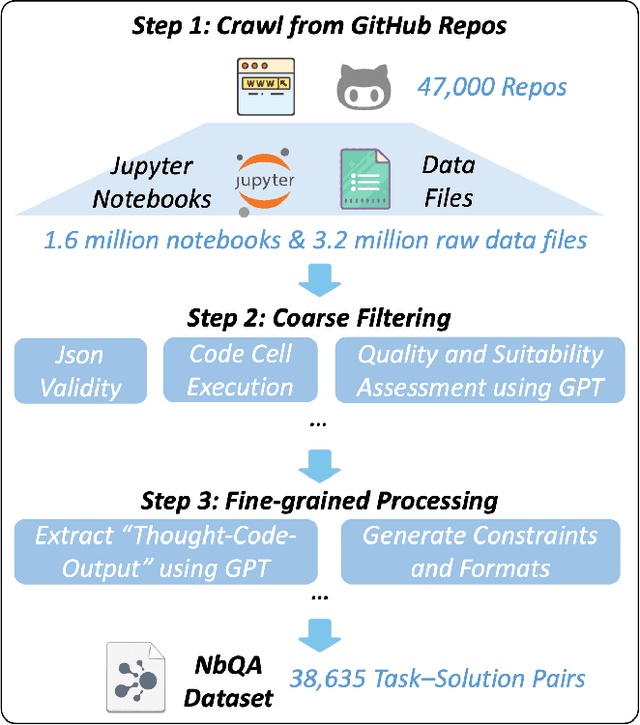
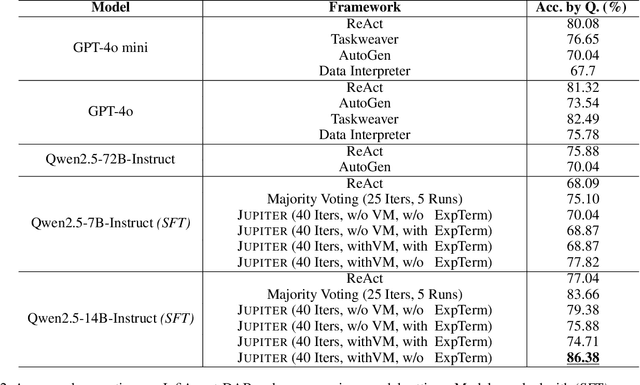
Abstract:Large language models (LLMs) have shown great promise in automating data science workflows, but existing models still struggle with multi-step reasoning and tool use, which limits their effectiveness on complex data analysis tasks. To address this, we propose a scalable pipeline that extracts high-quality, tool-based data analysis tasks and their executable multi-step solutions from real-world Jupyter notebooks and associated data files. Using this pipeline, we introduce NbQA, a large-scale dataset of standardized task-solution pairs that reflect authentic tool-use patterns in practical data science scenarios. To further enhance multi-step reasoning, we present Jupiter, a framework that formulates data analysis as a search problem and applies Monte Carlo Tree Search (MCTS) to generate diverse solution trajectories for value model learning. During inference, Jupiter combines the value model and node visit counts to efficiently collect executable multi-step plans with minimal search steps. Experimental results show that Qwen2.5-7B and 14B-Instruct models on NbQA solve 77.82% and 86.38% of tasks on InfiAgent-DABench, respectively-matching or surpassing GPT-4o and advanced agent frameworks. Further evaluations demonstrate improved generalization and stronger tool-use reasoning across diverse multi-step reasoning tasks.
Mem4Nav: Boosting Vision-and-Language Navigation in Urban Environments with a Hierarchical Spatial-Cognition Long-Short Memory System
Jun 24, 2025Abstract:Vision-and-Language Navigation (VLN) in large-scale urban environments requires embodied agents to ground linguistic instructions in complex scenes and recall relevant experiences over extended time horizons. Prior modular pipelines offer interpretability but lack unified memory, while end-to-end (M)LLM agents excel at fusing vision and language yet remain constrained by fixed context windows and implicit spatial reasoning. We introduce \textbf{Mem4Nav}, a hierarchical spatial-cognition long-short memory system that can augment any VLN backbone. Mem4Nav fuses a sparse octree for fine-grained voxel indexing with a semantic topology graph for high-level landmark connectivity, storing both in trainable memory tokens embedded via a reversible Transformer. Long-term memory (LTM) compresses and retains historical observations at both octree and graph nodes, while short-term memory (STM) caches recent multimodal entries in relative coordinates for real-time obstacle avoidance and local planning. At each step, STM retrieval sharply prunes dynamic context, and, when deeper history is needed, LTM tokens are decoded losslessly to reconstruct past embeddings. Evaluated on Touchdown and Map2Seq across three backbones (modular, state-of-the-art VLN with prompt-based LLM, and state-of-the-art VLN with strided-attention MLLM), Mem4Nav yields 7-13 pp gains in Task Completion, sufficient SPD reduction, and >10 pp nDTW improvement. Ablations confirm the indispensability of both the hierarchical map and dual memory modules. Our codes are open-sourced via https://github.com/tsinghua-fib-lab/Mem4Nav.
MRI-CORE: A Foundation Model for Magnetic Resonance Imaging
Jun 13, 2025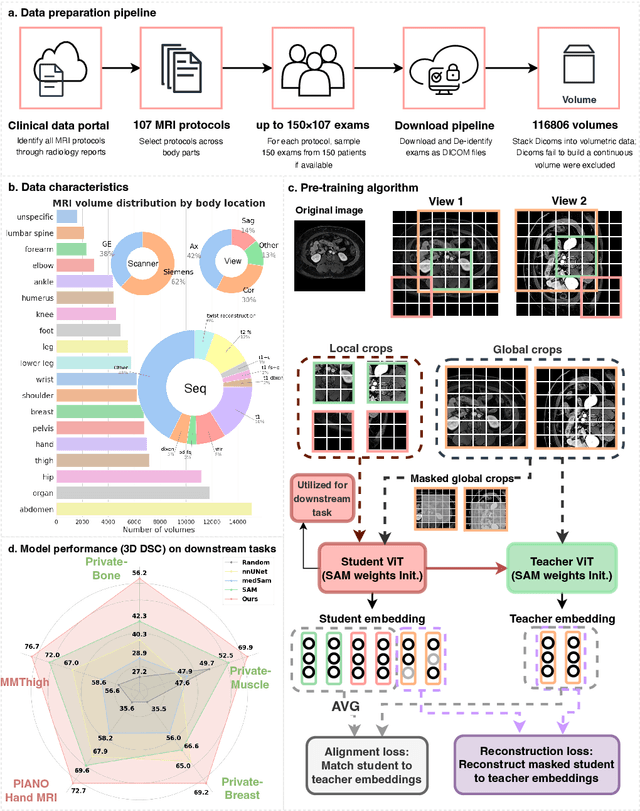
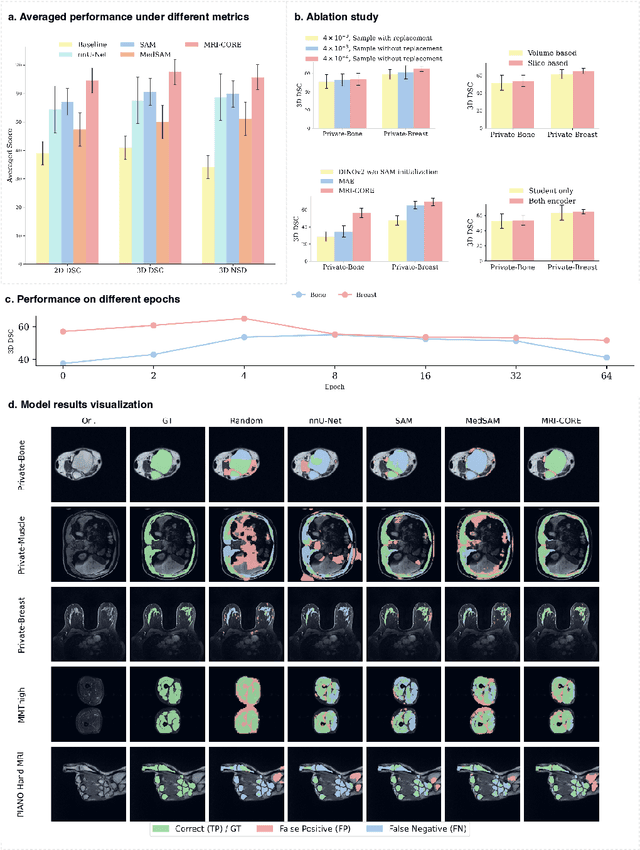
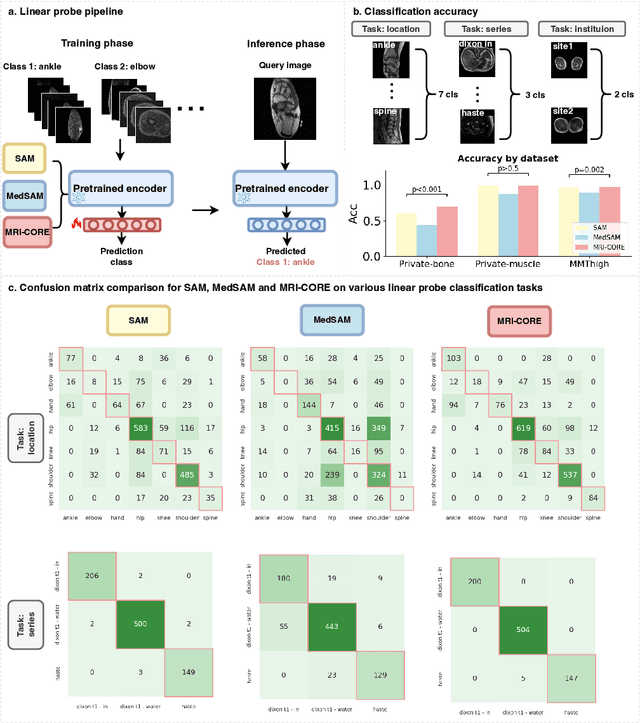
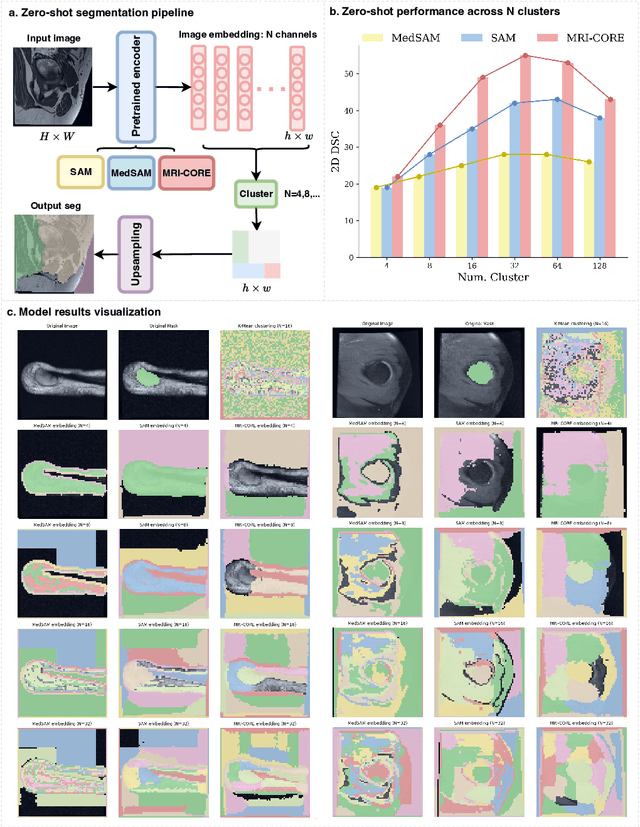
Abstract:The widespread use of Magnetic Resonance Imaging (MRI) and the rise of deep learning have enabled the development of powerful predictive models for a wide range of diagnostic tasks in MRI, such as image classification or object segmentation. However, training models for specific new tasks often requires large amounts of labeled data, which is difficult to obtain due to high annotation costs and data privacy concerns. To circumvent this issue, we introduce MRI-CORE (MRI COmprehensive Representation Encoder), a vision foundation model pre-trained using more than 6 million slices from over 110,000 MRI volumes across 18 main body locations. Experiments on five diverse object segmentation tasks in MRI demonstrate that MRI-CORE can significantly improve segmentation performance in realistic scenarios with limited labeled data availability, achieving an average gain of 6.97% 3D Dice Coefficient using only 10 annotated slices per task. We further demonstrate new model capabilities in MRI such as classification of image properties including body location, sequence type and institution, and zero-shot segmentation. These results highlight the value of MRI-CORE as a generalist vision foundation model for MRI, potentially lowering the data annotation resource barriers for many applications.
Bingo: Boosting Efficient Reasoning of LLMs via Dynamic and Significance-based Reinforcement Learning
Jun 09, 2025



Abstract:Large language models have demonstrated impressive reasoning capabilities, yet they often suffer from inefficiencies due to unnecessarily verbose or redundant outputs. While many works have explored reinforcement learning (RL) to enhance reasoning abilities, most primarily focus on improving accuracy, with limited attention to reasoning efficiency. Some existing approaches introduce direct length-based rewards to encourage brevity, but this often leads to noticeable drops in accuracy. In this paper, we propose Bingo, an RL framework that advances length-based reward design to boost efficient reasoning. Bingo incorporates two key mechanisms: a significance-aware length reward, which gradually guides the model to reduce only insignificant tokens, and a dynamic length reward, which initially encourages elaborate reasoning for hard questions but decays over time to improve overall efficiency. Experiments across multiple reasoning benchmarks show that Bingo improves both accuracy and efficiency. It outperforms the vanilla reward and several other length-based reward baselines in RL, achieving a favorable trade-off between accuracy and efficiency. These results underscore the potential of training LLMs explicitly for efficient reasoning.
MMTU: A Massive Multi-Task Table Understanding and Reasoning Benchmark
Jun 05, 2025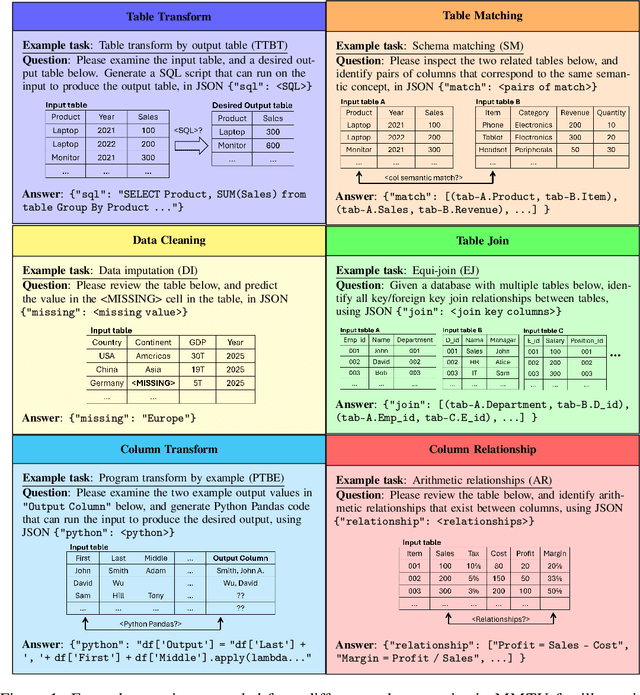
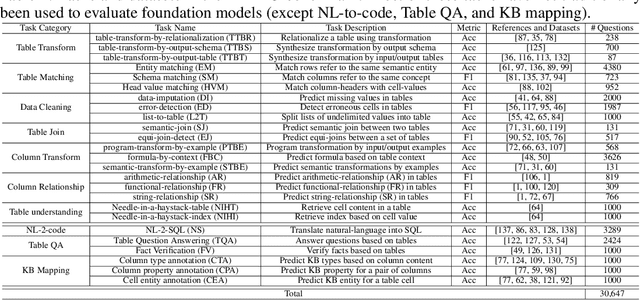
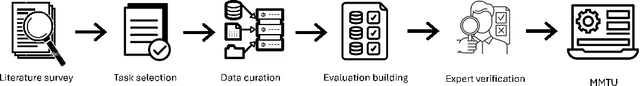
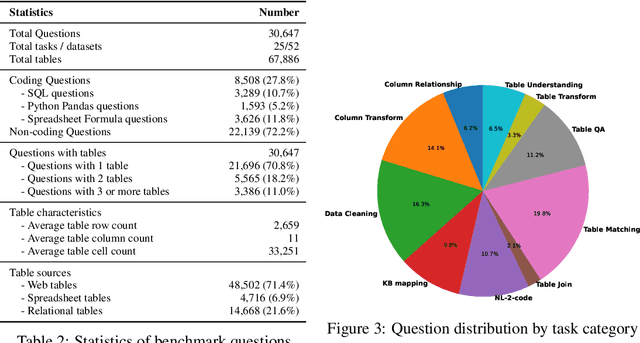
Abstract:Tables and table-based use cases play a crucial role in many important real-world applications, such as spreadsheets, databases, and computational notebooks, which traditionally require expert-level users like data engineers, data analysts, and database administrators to operate. Although LLMs have shown remarkable progress in working with tables (e.g., in spreadsheet and database copilot scenarios), comprehensive benchmarking of such capabilities remains limited. In contrast to an extensive and growing list of NLP benchmarks, evaluations of table-related tasks are scarce, and narrowly focus on tasks like NL-to-SQL and Table-QA, overlooking the broader spectrum of real-world tasks that professional users face. This gap limits our understanding and model progress in this important area. In this work, we introduce MMTU, a large-scale benchmark with over 30K questions across 25 real-world table tasks, designed to comprehensively evaluate models ability to understand, reason, and manipulate real tables at the expert-level. These tasks are drawn from decades' worth of computer science research on tabular data, with a focus on complex table tasks faced by professional users. We show that MMTU require a combination of skills -- including table understanding, reasoning, and coding -- that remain challenging for today's frontier models, where even frontier reasoning models like OpenAI o4-mini and DeepSeek R1 score only around 60%, suggesting significant room for improvement. We highlight key findings in our evaluation using MMTU and hope that this benchmark drives further advances in understanding and developing foundation models for structured data processing and analysis. Our code and data are available at https://github.com/MMTU-Benchmark/MMTU and https://huggingface.co/datasets/MMTU-benchmark/MMTU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge