Nicholas Konz
MRI-CORE: A Foundation Model for Magnetic Resonance Imaging
Jun 13, 2025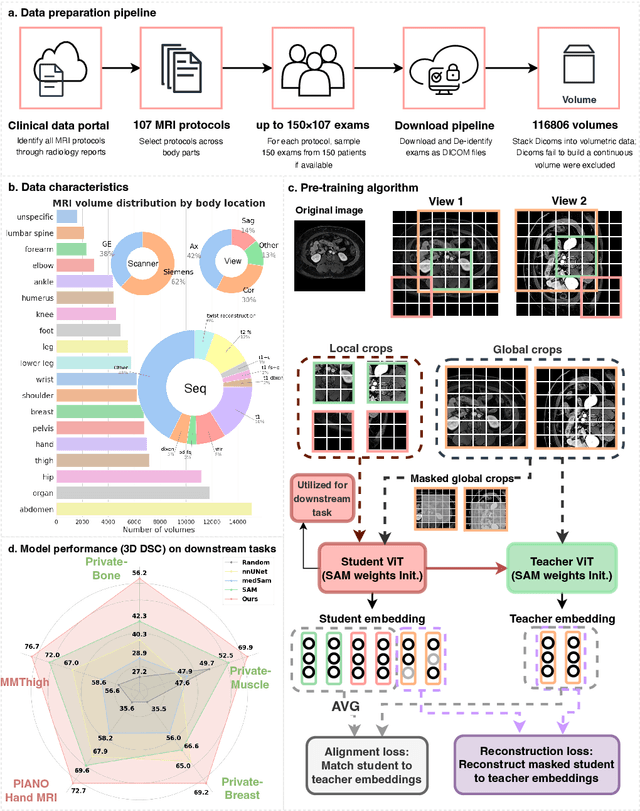
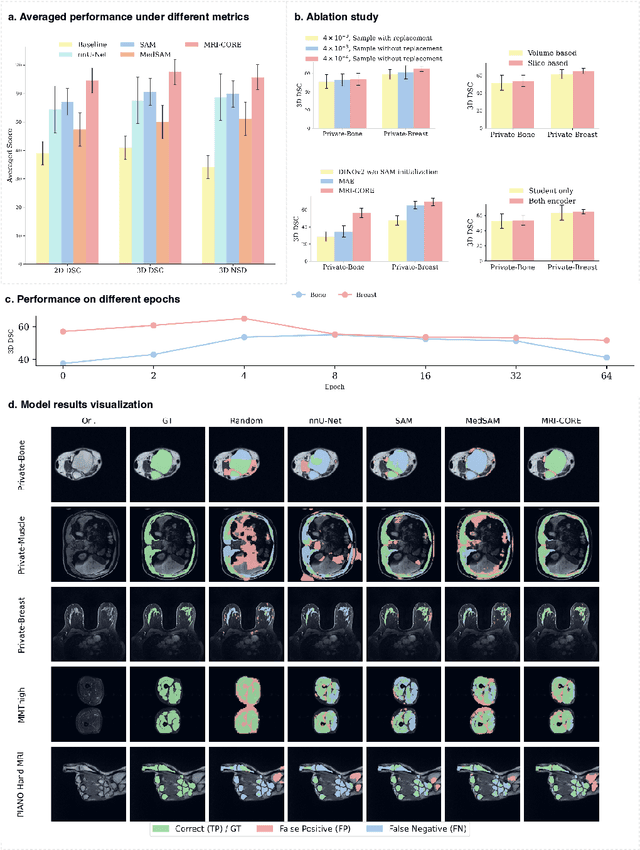
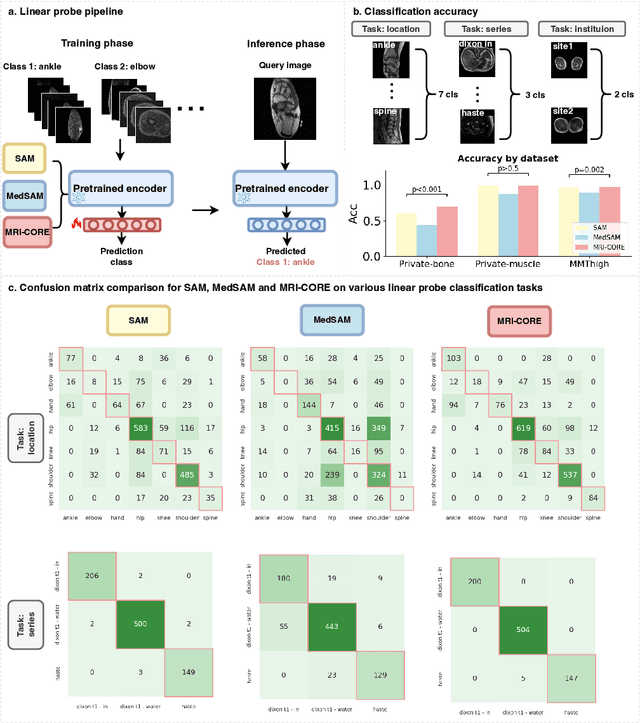
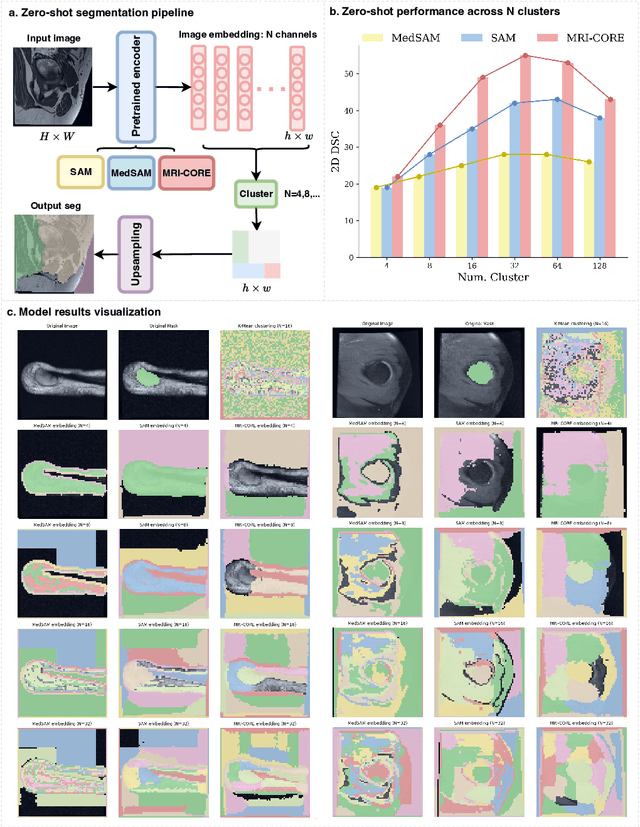
Abstract:The widespread use of Magnetic Resonance Imaging (MRI) and the rise of deep learning have enabled the development of powerful predictive models for a wide range of diagnostic tasks in MRI, such as image classification or object segmentation. However, training models for specific new tasks often requires large amounts of labeled data, which is difficult to obtain due to high annotation costs and data privacy concerns. To circumvent this issue, we introduce MRI-CORE (MRI COmprehensive Representation Encoder), a vision foundation model pre-trained using more than 6 million slices from over 110,000 MRI volumes across 18 main body locations. Experiments on five diverse object segmentation tasks in MRI demonstrate that MRI-CORE can significantly improve segmentation performance in realistic scenarios with limited labeled data availability, achieving an average gain of 6.97% 3D Dice Coefficient using only 10 annotated slices per task. We further demonstrate new model capabilities in MRI such as classification of image properties including body location, sequence type and institution, and zero-shot segmentation. These results highlight the value of MRI-CORE as a generalist vision foundation model for MRI, potentially lowering the data annotation resource barriers for many applications.
GuidedMorph: Two-Stage Deformable Registration for Breast MRI
May 19, 2025
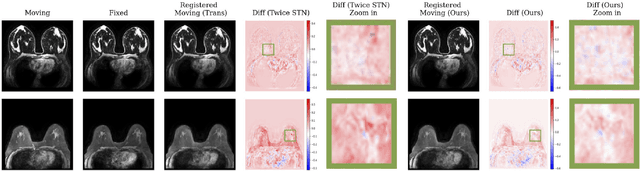
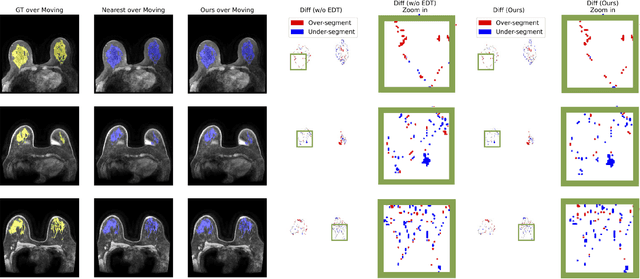
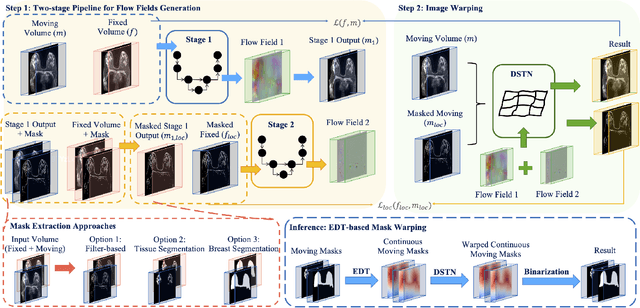
Abstract:Accurately registering breast MR images from different time points enables the alignment of anatomical structures and tracking of tumor progression, supporting more effective breast cancer detection, diagnosis, and treatment planning. However, the complexity of dense tissue and its highly non-rigid nature pose challenges for conventional registration methods, which primarily focus on aligning general structures while overlooking intricate internal details. To address this, we propose \textbf{GuidedMorph}, a novel two-stage registration framework designed to better align dense tissue. In addition to a single-scale network for global structure alignment, we introduce a framework that utilizes dense tissue information to track breast movement. The learned transformation fields are fused by introducing the Dual Spatial Transformer Network (DSTN), improving overall alignment accuracy. A novel warping method based on the Euclidean distance transform (EDT) is also proposed to accurately warp the registered dense tissue and breast masks, preserving fine structural details during deformation. The framework supports paradigms that require external segmentation models and with image data only. It also operates effectively with the VoxelMorph and TransMorph backbones, offering a versatile solution for breast registration. We validate our method on ISPY2 and internal dataset, demonstrating superior performance in dense tissue, overall breast alignment, and breast structural similarity index measure (SSIM), with notable improvements by over 13.01% in dense tissue Dice, 3.13% in breast Dice, and 1.21% in breast SSIM compared to the best learning-based baseline.
Accelerating Volumetric Medical Image Annotation via Short-Long Memory SAM 2
May 03, 2025Abstract:Manual annotation of volumetric medical images, such as magnetic resonance imaging (MRI) and computed tomography (CT), is a labor-intensive and time-consuming process. Recent advancements in foundation models for video object segmentation, such as Segment Anything Model 2 (SAM 2), offer a potential opportunity to significantly speed up the annotation process by manually annotating one or a few slices and then propagating target masks across the entire volume. However, the performance of SAM 2 in this context varies. Our experiments show that relying on a single memory bank and attention module is prone to error propagation, particularly at boundary regions where the target is present in the previous slice but absent in the current one. To address this problem, we propose Short-Long Memory SAM 2 (SLM-SAM 2), a novel architecture that integrates distinct short-term and long-term memory banks with separate attention modules to improve segmentation accuracy. We evaluate SLM-SAM 2 on three public datasets covering organs, bones, and muscles across MRI and CT modalities. We show that the proposed method markedly outperforms the default SAM 2, achieving average Dice Similarity Coefficient improvement of 0.14 and 0.11 in the scenarios when 5 volumes and 1 volume are available for the initial adaptation, respectively. SLM-SAM 2 also exhibits stronger resistance to over-propagation, making a notable step toward more accurate automated annotation of medical images for segmentation model development.
Quantifying the Limits of Segment Anything Model: Analyzing Challenges in Segmenting Tree-Like and Low-Contrast Structures
Dec 05, 2024Abstract:Segment Anything Model (SAM) has shown impressive performance in interactive and zero-shot segmentation across diverse domains, suggesting that they have learned a general concept of "objects" from their large-scale training. However, we observed that SAM struggles with certain types of objects, particularly those featuring dense, tree-like structures and low textural contrast from their surroundings. These failure modes are critical for understanding its limitations in real-world use. In order to systematically examine this issue, we propose metrics to quantify two key object characteristics: tree-likeness and textural separability. Through extensive controlled synthetic experiments and testing on real datasets, we demonstrate that SAM's performance is noticeably correlated with these factors. We link these behaviors under the concept of "textural confusion", where SAM misinterprets local structure as global texture, leading to over-segmentation, or struggles to differentiate objects from similarly textured backgrounds. These findings offer the first quantitative framework to model SAM's challenges, providing valuable insights into its limitations and guiding future improvements for vision foundation models.
RaD: A Metric for Medical Image Distribution Comparison in Out-of-Domain Detection and Other Applications
Dec 02, 2024Abstract:Determining whether two sets of images belong to the same or different domain is a crucial task in modern medical image analysis and deep learning, where domain shift is a common problem that commonly results in decreased model performance. This determination is also important to evaluate the output quality of generative models, e.g., image-to-image translation models used to mitigate domain shift. Current metrics for this either rely on the (potentially biased) choice of some downstream task such as segmentation, or adopt task-independent perceptual metrics (e.g., FID) from natural imaging which insufficiently capture anatomical consistency and realism in medical images. We introduce a new perceptual metric tailored for medical images: Radiomic Feature Distance (RaD), which utilizes standardized, clinically meaningful and interpretable image features. We show that RaD is superior to other metrics for out-of-domain (OOD) detection in a variety of experiments. Furthermore, RaD outperforms previous perceptual metrics (FID, KID, etc.) for image-to-image translation by correlating more strongly with downstream task performance as well as anatomical consistency and realism, and shows similar utility for evaluating unconditional image generation. RaD also offers additional benefits such as interpretability, as well as stability and computational efficiency at low sample sizes. Our results are supported by broad experiments spanning four multi-domain medical image datasets, nine downstream tasks, six image translation models, and other factors, highlighting the broad potential of RaD for medical image analysis.
The Impact of Scanner Domain Shift on Deep Learning Performance in Medical Imaging: an Experimental Study
Sep 06, 2024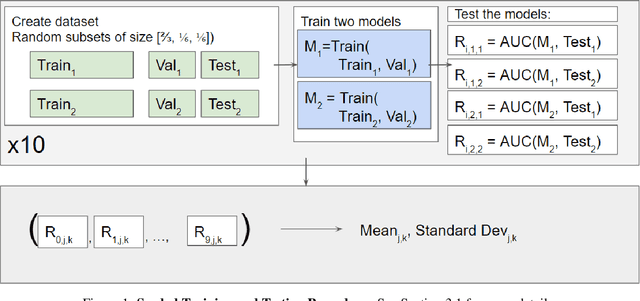
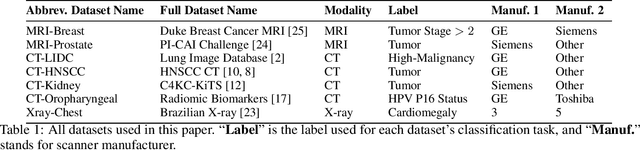
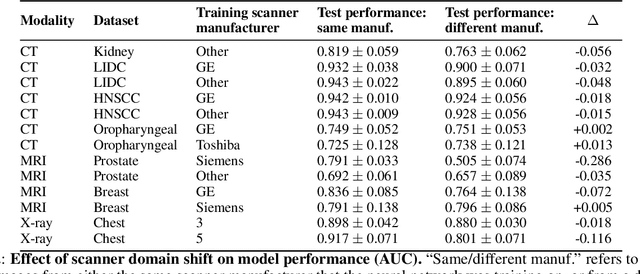
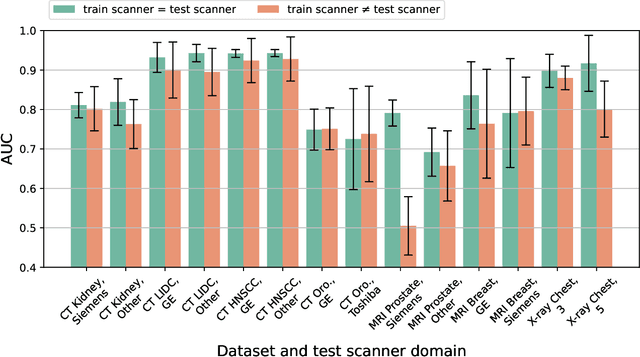
Abstract:Purpose: Medical images acquired using different scanners and protocols can differ substantially in their appearance. This phenomenon, scanner domain shift, can result in a drop in the performance of deep neural networks which are trained on data acquired by one scanner and tested on another. This significant practical issue is well-acknowledged, however, no systematic study of the issue is available across different modalities and diagnostic tasks. Materials and Methods: In this paper, we present a broad experimental study evaluating the impact of scanner domain shift on convolutional neural network performance for different automated diagnostic tasks. We evaluate this phenomenon in common radiological modalities, including X-ray, CT, and MRI. Results: We find that network performance on data from a different scanner is almost always worse than on same-scanner data, and we quantify the degree of performance drop across different datasets. Notably, we find that this drop is most severe for MRI, moderate for X-ray, and quite small for CT, on average, which we attribute to the standardized nature of CT acquisition systems which is not present in MRI or X-ray. We also study how injecting varying amounts of target domain data into the training set, as well as adding noise to the training data, helps with generalization. Conclusion: Our results provide extensive experimental evidence and quantification of the extent of performance drop caused by scanner domain shift in deep learning across different modalities, with the goal of guiding the future development of robust deep learning models for medical image analysis.
Pre-processing and Compression: Understanding Hidden Representation Refinement Across Imaging Domains via Intrinsic Dimension
Aug 15, 2024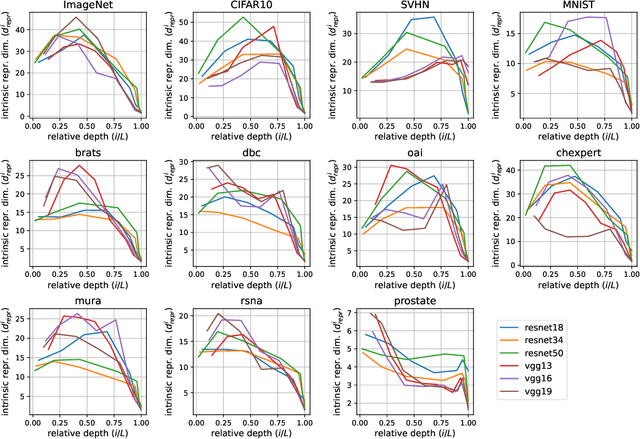

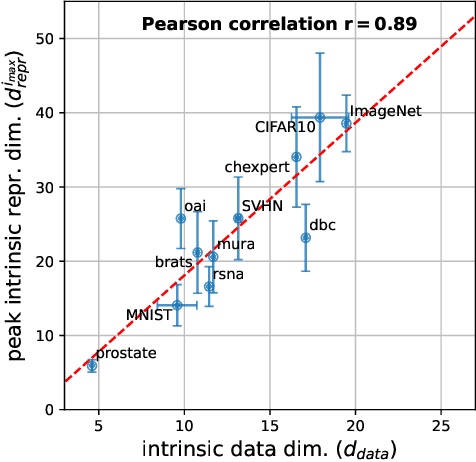
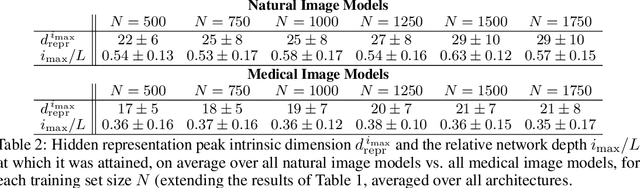
Abstract:In recent years, there has been interest in how geometric properties such as intrinsic dimension (ID) of a neural network's hidden representations evolve through its layers, and how such properties are predictive of important model behavior such as generalization ability. However, evidence has begun to emerge that such behavior can change significantly depending on the domain of the network's training data, such as natural versus medical images. Here, we further this inquiry by exploring how the ID of a network's learned representations evolves through its layers, in essence, characterizing how the network successively refines the information content of input data to be used for predictions. Analyzing eleven natural and medical image datasets across six network architectures, we find that the shape of this ID evolution curve differs noticeably between natural and medical image models: medical image models peak in representation ID earlier in the network, implying a difference in the image features and their abstractness that are typically used for downstream tasks in these domains. Additionally, we discover a strong correlation of this peak representation ID with the ID of the data in its input space, implying that the intrinsic information content of a model's learned representations is guided by that of the data it was trained on. Overall, our findings emphasize notable discrepancies in network behavior between natural and non-natural imaging domains regarding hidden representation information content, and provide further insights into how a network's learned features are shaped by its training data.
Rethinking Perceptual Metrics for Medical Image Translation
Apr 10, 2024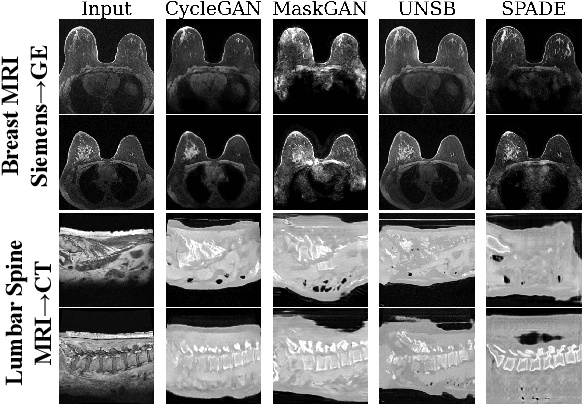
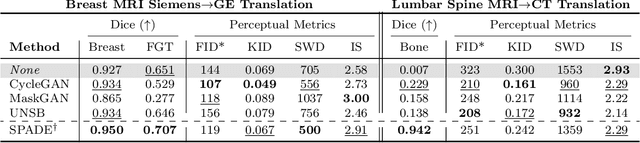
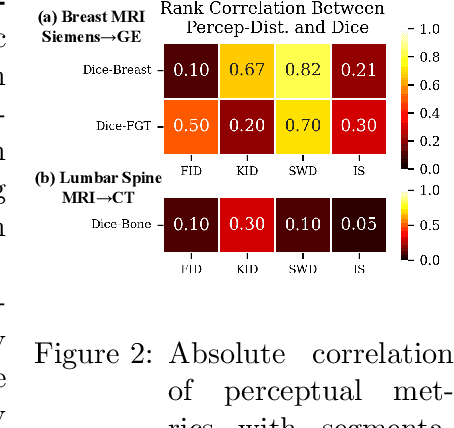
Abstract:Modern medical image translation methods use generative models for tasks such as the conversion of CT images to MRI. Evaluating these methods typically relies on some chosen downstream task in the target domain, such as segmentation. On the other hand, task-agnostic metrics are attractive, such as the network feature-based perceptual metrics (e.g., FID) that are common to image translation in general computer vision. In this paper, we investigate evaluation metrics for medical image translation on two medical image translation tasks (GE breast MRI to Siemens breast MRI and lumbar spine MRI to CT), tested on various state-of-the-art translation methods. We show that perceptual metrics do not generally correlate with segmentation metrics due to them extending poorly to the anatomical constraints of this sub-field, with FID being especially inconsistent. However, we find that the lesser-used pixel-level SWD metric may be useful for subtle intra-modality translation. Our results demonstrate the need for further research into helpful metrics for medical image translation.
ContourDiff: Unpaired Image Translation with Contour-Guided Diffusion Models
Mar 16, 2024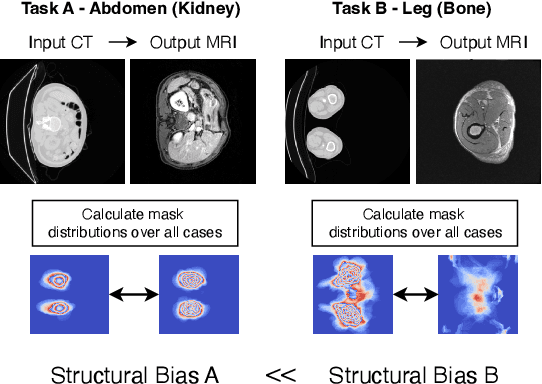
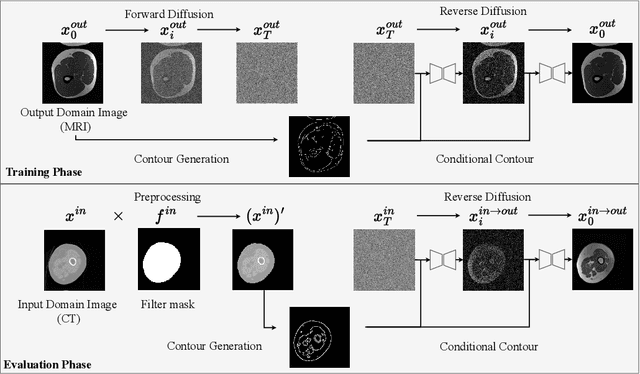
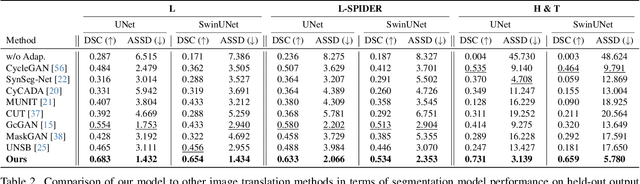
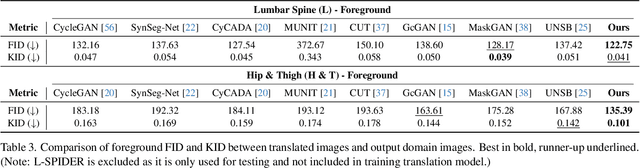
Abstract:Accurately translating medical images across different modalities (e.g., CT to MRI) has numerous downstream clinical and machine learning applications. While several methods have been proposed to achieve this, they often prioritize perceptual quality with respect to output domain features over preserving anatomical fidelity. However, maintaining anatomy during translation is essential for many tasks, e.g., when leveraging masks from the input domain to develop a segmentation model with images translated to the output domain. To address these challenges, we propose ContourDiff, a novel framework that leverages domain-invariant anatomical contour representations of images. These representations are simple to extract from images, yet form precise spatial constraints on their anatomical content. We introduce a diffusion model that converts contour representations of images from arbitrary input domains into images in the output domain of interest. By applying the contour as a constraint at every diffusion sampling step, we ensure the preservation of anatomical content. We evaluate our method by training a segmentation model on images translated from CT to MRI with their original CT masks and testing its performance on real MRIs. Our method outperforms other unpaired image translation methods by a significant margin, furthermore without the need to access any input domain information during training.
Medical Image Segmentation with InTEnt: Integrated Entropy Weighting for Single Image Test-Time Adaptation
Feb 16, 2024



Abstract:Test-time adaptation (TTA) refers to adapting a trained model to a new domain during testing. Existing TTA techniques rely on having multiple test images from the same domain, yet this may be impractical in real-world applications such as medical imaging, where data acquisition is expensive and imaging conditions vary frequently. Here, we approach such a task, of adapting a medical image segmentation model with only a single unlabeled test image. Most TTA approaches, which directly minimize the entropy of predictions, fail to improve performance significantly in this setting, in which we also observe the choice of batch normalization (BN) layer statistics to be a highly important yet unstable factor due to only having a single test domain example. To overcome this, we propose to instead integrate over predictions made with various estimates of target domain statistics between the training and test statistics, weighted based on their entropy statistics. Our method, validated on 24 source/target domain splits across 3 medical image datasets surpasses the leading method by 2.9% Dice coefficient on average.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge