Maciej A. Mazurowski
Diagnostic Impact of Cine Clips for Thyroid Nodule Assessment on Ultrasound
Feb 01, 2026Abstract:Background: Thyroid ultrasound is commonly performed using a combination of static images and cine clips (video recordings). However, the exact utility and impact of cine images remains unknown. This study aimed to evaluate the impact of cine imaging on accuracy and consistency of thyroid nodule assessment, using the American College of Radiology Thyroid Reporting and Data System (ACR TI-RADS). Methods: 50 benign and 50 malignant thyroid nodules with cytopathology results were included. A reader study with 4 specialty-trained radiologists was then conducted over 3 rounds, assessing only static images in the first two rounds and both static and cine images in the third round. TI-RADS scores and the consequent management recommendations were then evaluated by comparing them to the malignancy status of the nodules. Results: Mean sensitivity for malignancy detection was 0.65 for static images and 0.67 with both static and cine images (p>0.5). Specificity was 0.20 for static images and 0.22 with both static and cine images (p>0.5). Management recommendations were similar with and without cine images. Intrareader agreement on feature assignments remained consistent across all rounds, though TI-RADS point totals were slightly higher with cine images. Conclusion: The inclusion of cine imaging for thyroid nodule assessment on ultrasound did not significantly change diagnostic performance. Current practice guidelines, which do not mandate cine imaging, are sufficient for accurate diagnosis.
Effectiveness of Automatically Curated Dataset in Thyroid Nodules Classification Algorithms Using Deep Learning
Feb 01, 2026Abstract:The diagnosis of thyroid nodule cancers commonly utilizes ultrasound images. Several studies showed that deep learning algorithms designed to classify benign and malignant thyroid nodules could match radiologists' performance. However, data availability for training deep learning models is often limited due to the significant effort required to curate such datasets. The previous study proposed a method to curate thyroid nodule datasets automatically. It was tested to have a 63% yield rate and 83% accuracy. However, the usefulness of the generated data for training deep learning models remains unknown. In this study, we conducted experiments to determine whether using a automatically-curated dataset improves deep learning algorithms' performance. We trained deep learning models on the manually annotated and automatically-curated datasets. We also trained with a smaller subset of the automatically-curated dataset that has higher accuracy to explore the optimum usage of such dataset. As a result, the deep learning model trained on the manually selected dataset has an AUC of 0.643 (95% confidence interval [CI]: 0.62, 0.66). It is significantly lower than the AUC of the 6automatically-curated dataset trained deep learning model, 0.694 (95% confidence interval [CI]: 0.67, 0.73, P < .001). The AUC of the accurate subset trained deep learning model is 0.689 (95% confidence interval [CI]: 0.66, 0.72, P > .43), which is insignificantly worse than the AUC of the full automatically-curated dataset. In conclusion, we showed that using a automatically-curated dataset can substantially increase the performance of deep learning algorithms, and it is suggested to use all the data rather than only using the accurate subset.
Automated glenoid bone loss measurement and segmentation in CT scans for pre-operative planning in shoulder instability
Nov 18, 2025Abstract:Reliable measurement of glenoid bone loss is essential for operative planning in shoulder instability, but current manual and semi-automated methods are time-consuming and often subject to interreader variability. We developed and validated a fully automated deep learning pipeline for measuring glenoid bone loss on three-dimensional computed tomography (CT) scans using a linear-based, en-face view, best-circle method. Shoulder CT images of 91 patients (average age, 40 years; range, 14-89 years; 65 men) were retrospectively collected along with manual labels including glenoid segmentation, landmarks, and bone loss measurements. The multi-stage algorithm has three main stages: (1) segmentation, where we developed a U-Net to automatically segment the glenoid and humerus; (2) anatomical landmark detection, where a second network predicts glenoid rim points; and (3) geometric fitting, where we applied principal component analysis (PCA), projection, and circle fitting to compute the percentage of bone loss. The automated measurements showed strong agreement with consensus readings and exceeded surgeon-to-surgeon consistency (intraclass correlation coefficient (ICC) 0.84 vs 0.78), including in low- and high-bone-loss subgroups (ICC 0.71 vs 0.63 and 0.83 vs 0.21, respectively; P < 0.001). For classifying patients into low, medium, and high bone-loss categories, the pipeline achieved a recall of 0.714 for low and 0.857 for high severity, with no low cases misclassified as high or vice versa. These results suggest that our method is a time-efficient and clinically reliable tool for preoperative planning in shoulder instability and for screening patients with substantial glenoid bone loss. Code and dataset are available at https://github.com/Edenliu1/Auto-Glenoid-Measurement-DL-Pipeline.
Transplant-Ready? Evaluating AI Lung Segmentation Models in Candidates with Severe Lung Disease
Sep 18, 2025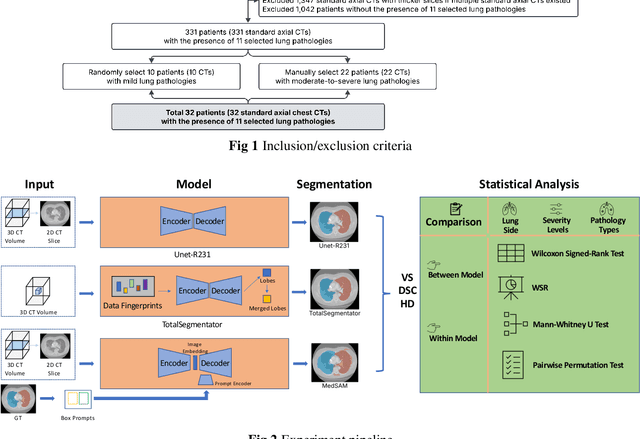
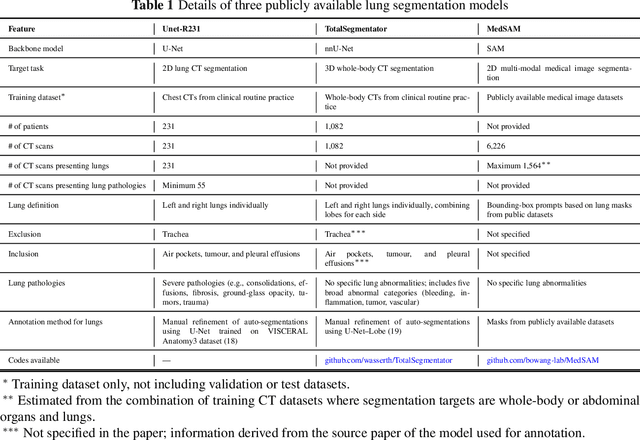
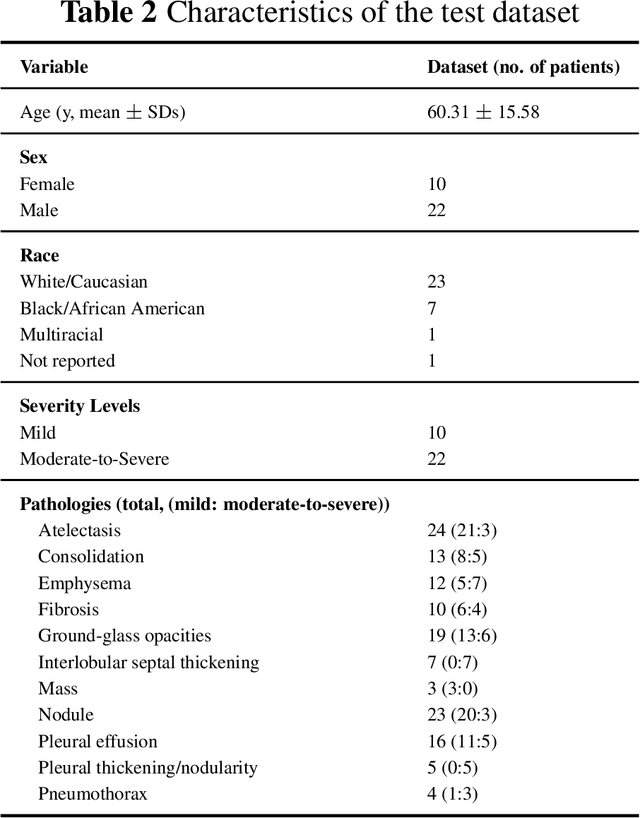
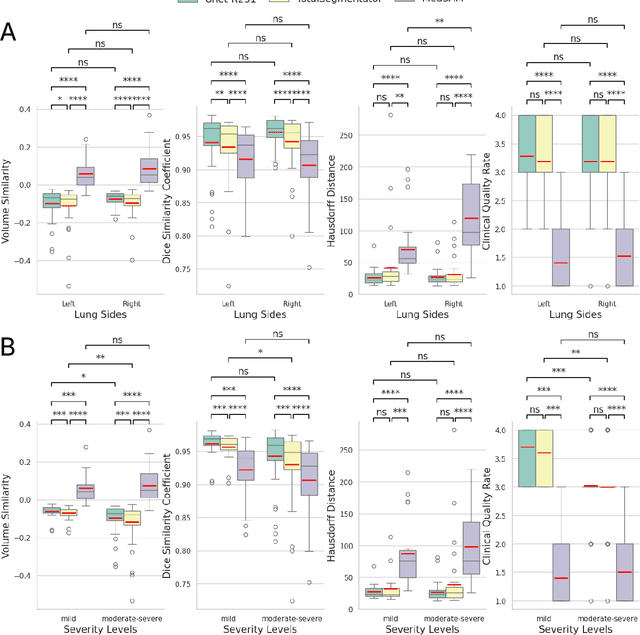
Abstract:This study evaluates publicly available deep-learning based lung segmentation models in transplant-eligible patients to determine their performance across disease severity levels, pathology categories, and lung sides, and to identify limitations impacting their use in preoperative planning in lung transplantation. This retrospective study included 32 patients who underwent chest CT scans at Duke University Health System between 2017 and 2019 (total of 3,645 2D axial slices). Patients with standard axial CT scans were selected based on the presence of two or more lung pathologies of varying severity. Lung segmentation was performed using three previously developed deep learning models: Unet-R231, TotalSegmentator, MedSAM. Performance was assessed using quantitative metrics (volumetric similarity, Dice similarity coefficient, Hausdorff distance) and a qualitative measure (four-point clinical acceptability scale). Unet-R231 consistently outperformed TotalSegmentator and MedSAM in general, for different severity levels, and pathology categories (p<0.05). All models showed significant performance declines from mild to moderate-to-severe cases, particularly in volumetric similarity (p<0.05), without significant differences among lung sides or pathology types. Unet-R231 provided the most accurate automated lung segmentation among evaluated models with TotalSegmentator being a close second, though their performance declined significantly in moderate-to-severe cases, emphasizing the need for specialized model fine-tuning in severe pathology contexts.
BreastSegNet: Multi-label Segmentation of Breast MRI
Jul 18, 2025Abstract:Breast MRI provides high-resolution imaging critical for breast cancer screening and preoperative staging. However, existing segmentation methods for breast MRI remain limited in scope, often focusing on only a few anatomical structures, such as fibroglandular tissue or tumors, and do not cover the full range of tissues seen in scans. This narrows their utility for quantitative analysis. In this study, we present BreastSegNet, a multi-label segmentation algorithm for breast MRI that covers nine anatomical labels: fibroglandular tissue (FGT), vessel, muscle, bone, lesion, lymph node, heart, liver, and implant. We manually annotated a large set of 1123 MRI slices capturing these structures with detailed review and correction from an expert radiologist. Additionally, we benchmark nine segmentation models, including U-Net, SwinUNet, UNet++, SAM, MedSAM, and nnU-Net with multiple ResNet-based encoders. Among them, nnU-Net ResEncM achieves the highest average Dice scores of 0.694 across all labels. It performs especially well on heart, liver, muscle, FGT, and bone, with Dice scores exceeding 0.73, and approaching 0.90 for heart and liver. All model code and weights are publicly available, and we plan to release the data at a later date.
Improving Surgical Risk Prediction Through Integrating Automated Body Composition Analysis: a Retrospective Trial on Colectomy Surgery
Jun 16, 2025Abstract:Objective: To evaluate whether preoperative body composition metrics automatically extracted from CT scans can predict postoperative outcomes after colectomy, either alone or combined with clinical variables or existing risk predictors. Main outcomes and measures: The primary outcome was the predictive performance for 1-year all-cause mortality following colectomy. A Cox proportional hazards model with 1-year follow-up was used, and performance was evaluated using the concordance index (C-index) and Integrated Brier Score (IBS). Secondary outcomes included postoperative complications, unplanned readmission, blood transfusion, and severe infection, assessed using AUC and Brier Score from logistic regression. Odds ratios (OR) described associations between individual CT-derived body composition metrics and outcomes. Over 300 features were extracted from preoperative CTs across multiple vertebral levels, including skeletal muscle area, density, fat areas, and inter-tissue metrics. NSQIP scores were available for all surgeries after 2012.
MRI-CORE: A Foundation Model for Magnetic Resonance Imaging
Jun 13, 2025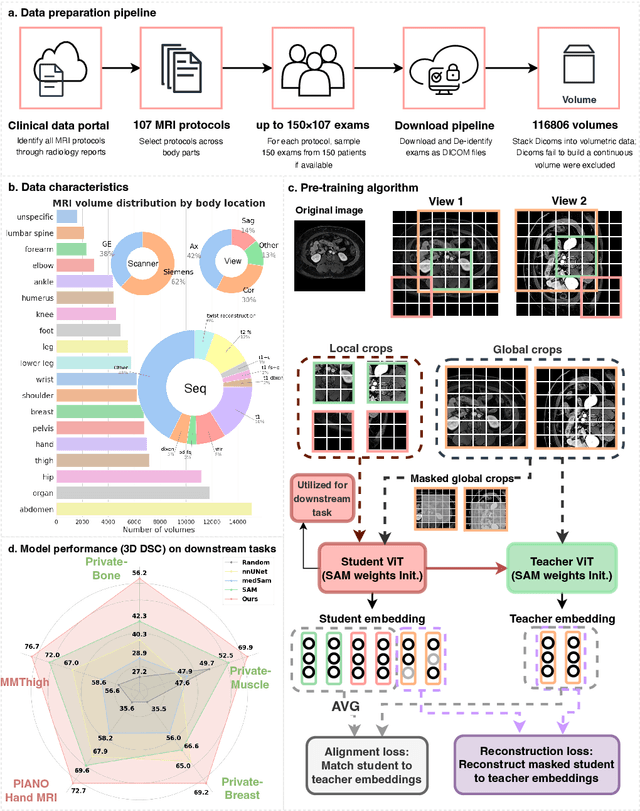
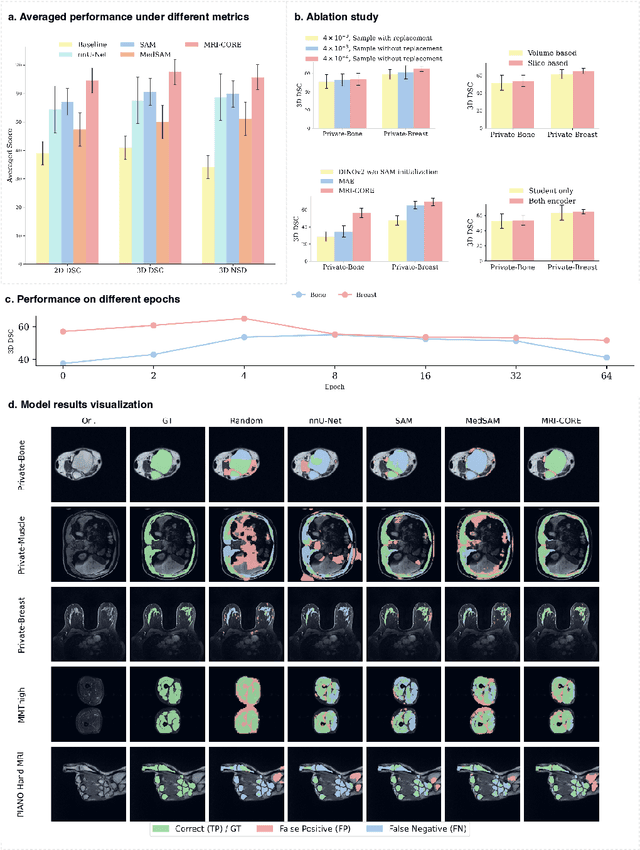
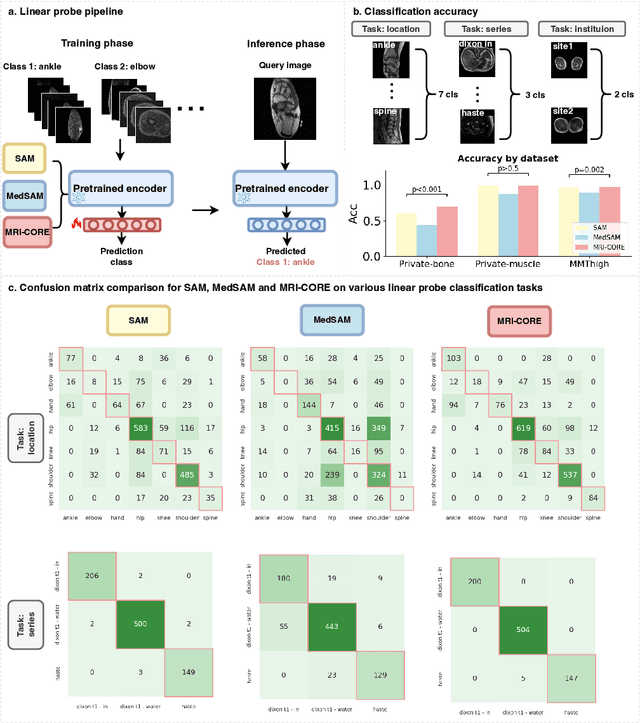
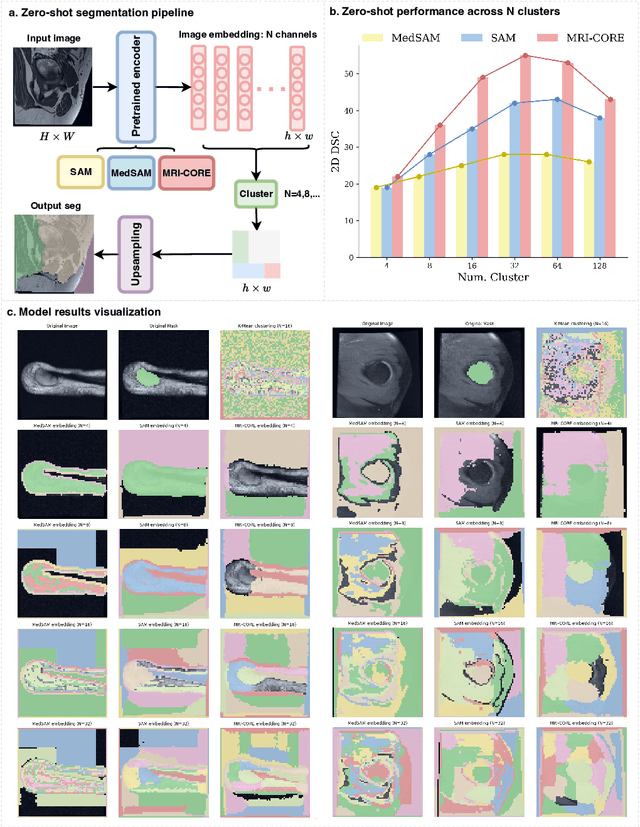
Abstract:The widespread use of Magnetic Resonance Imaging (MRI) and the rise of deep learning have enabled the development of powerful predictive models for a wide range of diagnostic tasks in MRI, such as image classification or object segmentation. However, training models for specific new tasks often requires large amounts of labeled data, which is difficult to obtain due to high annotation costs and data privacy concerns. To circumvent this issue, we introduce MRI-CORE (MRI COmprehensive Representation Encoder), a vision foundation model pre-trained using more than 6 million slices from over 110,000 MRI volumes across 18 main body locations. Experiments on five diverse object segmentation tasks in MRI demonstrate that MRI-CORE can significantly improve segmentation performance in realistic scenarios with limited labeled data availability, achieving an average gain of 6.97% 3D Dice Coefficient using only 10 annotated slices per task. We further demonstrate new model capabilities in MRI such as classification of image properties including body location, sequence type and institution, and zero-shot segmentation. These results highlight the value of MRI-CORE as a generalist vision foundation model for MRI, potentially lowering the data annotation resource barriers for many applications.
GuidedMorph: Two-Stage Deformable Registration for Breast MRI
May 19, 2025
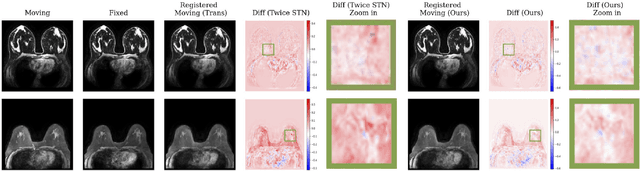
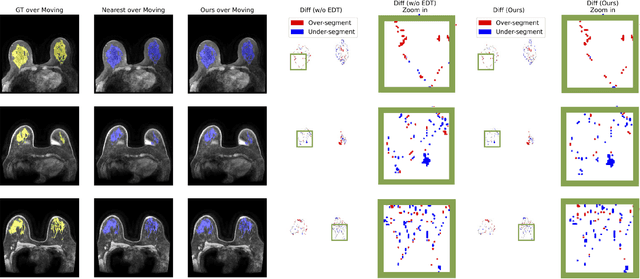
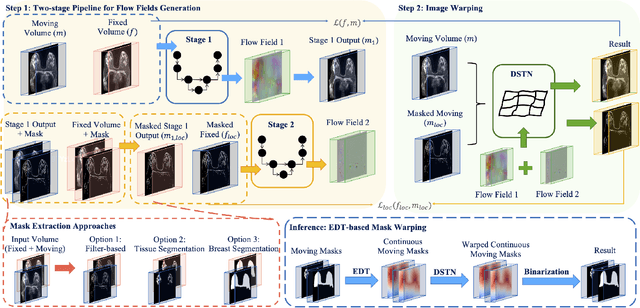
Abstract:Accurately registering breast MR images from different time points enables the alignment of anatomical structures and tracking of tumor progression, supporting more effective breast cancer detection, diagnosis, and treatment planning. However, the complexity of dense tissue and its highly non-rigid nature pose challenges for conventional registration methods, which primarily focus on aligning general structures while overlooking intricate internal details. To address this, we propose \textbf{GuidedMorph}, a novel two-stage registration framework designed to better align dense tissue. In addition to a single-scale network for global structure alignment, we introduce a framework that utilizes dense tissue information to track breast movement. The learned transformation fields are fused by introducing the Dual Spatial Transformer Network (DSTN), improving overall alignment accuracy. A novel warping method based on the Euclidean distance transform (EDT) is also proposed to accurately warp the registered dense tissue and breast masks, preserving fine structural details during deformation. The framework supports paradigms that require external segmentation models and with image data only. It also operates effectively with the VoxelMorph and TransMorph backbones, offering a versatile solution for breast registration. We validate our method on ISPY2 and internal dataset, demonstrating superior performance in dense tissue, overall breast alignment, and breast structural similarity index measure (SSIM), with notable improvements by over 13.01% in dense tissue Dice, 3.13% in breast Dice, and 1.21% in breast SSIM compared to the best learning-based baseline.
Accelerating Volumetric Medical Image Annotation via Short-Long Memory SAM 2
May 03, 2025Abstract:Manual annotation of volumetric medical images, such as magnetic resonance imaging (MRI) and computed tomography (CT), is a labor-intensive and time-consuming process. Recent advancements in foundation models for video object segmentation, such as Segment Anything Model 2 (SAM 2), offer a potential opportunity to significantly speed up the annotation process by manually annotating one or a few slices and then propagating target masks across the entire volume. However, the performance of SAM 2 in this context varies. Our experiments show that relying on a single memory bank and attention module is prone to error propagation, particularly at boundary regions where the target is present in the previous slice but absent in the current one. To address this problem, we propose Short-Long Memory SAM 2 (SLM-SAM 2), a novel architecture that integrates distinct short-term and long-term memory banks with separate attention modules to improve segmentation accuracy. We evaluate SLM-SAM 2 on three public datasets covering organs, bones, and muscles across MRI and CT modalities. We show that the proposed method markedly outperforms the default SAM 2, achieving average Dice Similarity Coefficient improvement of 0.14 and 0.11 in the scenarios when 5 volumes and 1 volume are available for the initial adaptation, respectively. SLM-SAM 2 also exhibits stronger resistance to over-propagation, making a notable step toward more accurate automated annotation of medical images for segmentation model development.
Breast density in MRI: an AI-based quantification and relationship to assessment in mammography
Apr 21, 2025Abstract:Mammographic breast density is a well-established risk factor for breast cancer. Recently there has been interest in breast MRI as an adjunct to mammography, as this modality provides an orthogonal and highly quantitative assessment of breast tissue. However, its 3D nature poses analytic challenges related to delineating and aggregating complex structures across slices. Here, we applied an in-house machine-learning algorithm to assess breast density on normal breasts in three MRI datasets. Breast density was consistent across different datasets (0.104 - 0.114). Analysis across different age groups also demonstrated strong consistency across datasets and confirmed a trend of decreasing density with age as reported in previous studies. MR breast density was correlated with mammographic breast density, although some notable differences suggest that certain breast density components are captured only on MRI. Future work will determine how to integrate MR breast density with current tools to improve future breast cancer risk prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge