Maximilian Rokuss
CRONOS: Continuous Time Reconstruction for 4D Medical Longitudinal Series
Dec 18, 2025Abstract:Forecasting how 3D medical scans evolve over time is important for disease progression, treatment planning, and developmental assessment. Yet existing models either rely on a single prior scan, fixed grid times, or target global labels, which limits voxel-level forecasting under irregular sampling. We present CRONOS, a unified framework for many-to-one prediction from multiple past scans that supports both discrete (grid-based) and continuous (real-valued) timestamps in one model, to the best of our knowledge the first to achieve continuous sequence-to-image forecasting for 3D medical data. CRONOS learns a spatio-temporal velocity field that transports context volumes toward a target volume at an arbitrary time, while operating directly in 3D voxel space. Across three public datasets spanning Cine-MRI, perfusion CT, and longitudinal MRI, CRONOS outperforms other baselines, while remaining computationally competitive. We will release code and evaluation protocols to enable reproducible, multi-dataset benchmarking of multi-context, continuous-time forecasting.
VoxTell: Free-Text Promptable Universal 3D Medical Image Segmentation
Nov 14, 2025Abstract:We introduce VoxTell, a vision-language model for text-prompted volumetric medical image segmentation. It maps free-form descriptions, from single words to full clinical sentences, to 3D masks. Trained on 62K+ CT, MRI, and PET volumes spanning over 1K anatomical and pathological classes, VoxTell uses multi-stage vision-language fusion across decoder layers to align textual and visual features at multiple scales. It achieves state-of-the-art zero-shot performance across modalities on unseen datasets, excelling on familiar concepts while generalizing to related unseen classes. Extensive experiments further demonstrate strong cross-modality transfer, robustness to linguistic variations and clinical language, as well as accurate instance-specific segmentation from real-world text. Code is available at: https://www.github.com/MIC-DKFZ/VoxTell
MeisenMeister: A Simple Two Stage Pipeline for Breast Cancer Classification on MRI
Oct 31, 2025Abstract:The ODELIA Breast MRI Challenge 2025 addresses a critical issue in breast cancer screening: improving early detection through more efficient and accurate interpretation of breast MRI scans. Even though methods for general-purpose whole-body lesion segmentation as well as multi-time-point analysis exist, breast cancer detection remains highly challenging, largely due to the limited availability of high-quality segmentation labels. Therefore, developing robust classification-based approaches is crucial for the future of early breast cancer detection, particularly in applications such as large-scale screening. In this write-up, we provide a comprehensive overview of our approach to the challenge. We begin by detailing the underlying concept and foundational assumptions that guided our work. We then describe the iterative development process, highlighting the key stages of experimentation, evaluation, and refinement that shaped the evolution of our solution. Finally, we present the reasoning and evidence that informed the design choices behind our final submission, with a focus on performance, robustness, and clinical relevance. We release our full implementation publicly at https://github.com/MIC-DKFZ/MeisenMeister
Towards Interactive Lesion Segmentation in Whole-Body PET/CT with Promptable Models
Aug 29, 2025Abstract:Whole-body PET/CT is a cornerstone of oncological imaging, yet accurate lesion segmentation remains challenging due to tracer heterogeneity, physiological uptake, and multi-center variability. While fully automated methods have advanced substantially, clinical practice benefits from approaches that keep humans in the loop to efficiently refine predicted masks. The autoPET/CT IV challenge addresses this need by introducing interactive segmentation tasks based on simulated user prompts. In this work, we present our submission to Task 1. Building on the winning autoPET III nnU-Net pipeline, we extend the framework with promptable capabilities by encoding user-provided foreground and background clicks as additional input channels. We systematically investigate representations for spatial prompts and demonstrate that Euclidean Distance Transform (EDT) encodings consistently outperform Gaussian kernels. Furthermore, we propose online simulation of user interactions and a custom point sampling strategy to improve robustness under realistic prompting conditions. Our ensemble of EDT-based models, trained with and without external data, achieves the strongest cross-validation performance, reducing both false positives and false negatives compared to baseline models. These results highlight the potential of promptable models to enable efficient, user-guided segmentation workflows in multi-tracer, multi-center PET/CT. Code is publicly available at https://github.com/MIC-DKFZ/autoPET-interactive
A Multi-Stage Fine-Tuning and Ensembling Strategy for Pancreatic Tumor Segmentation in Diagnostic and Therapeutic MRI
Aug 29, 2025Abstract:Automated segmentation of Pancreatic Ductal Adenocarcinoma (PDAC) from MRI is critical for clinical workflows but is hindered by poor tumor-tissue contrast and a scarcity of annotated data. This paper details our submission to the PANTHER challenge, addressing both diagnostic T1-weighted (Task 1) and therapeutic T2-weighted (Task 2) segmentation. Our approach is built upon the nnU-Net framework and leverages a deep, multi-stage cascaded pre-training strategy, starting from a general anatomical foundation model and sequentially fine-tuning on CT pancreatic lesion datasets and the target MRI modalities. Through extensive five-fold cross-validation, we systematically evaluated data augmentation schemes and training schedules. Our analysis revealed a critical trade-off, where aggressive data augmentation produced the highest volumetric accuracy, while default augmentations yielded superior boundary precision (achieving a state-of-the-art MASD of 5.46 mm and HD95 of 17.33 mm for Task 1). For our final submission, we exploited this finding by constructing custom, heterogeneous ensembles of specialist models, essentially creating a mix of experts. This metric-aware ensembling strategy proved highly effective, achieving a top cross-validation Tumor Dice score of 0.661 for Task 1 and 0.523 for Task 2. Our work presents a robust methodology for developing specialized, high-performance models in the context of limited data and complex medical imaging tasks (Team MIC-DKFZ).
Temporal Flow Matching for Learning Spatio-Temporal Trajectories in 4D Longitudinal Medical Imaging
Aug 29, 2025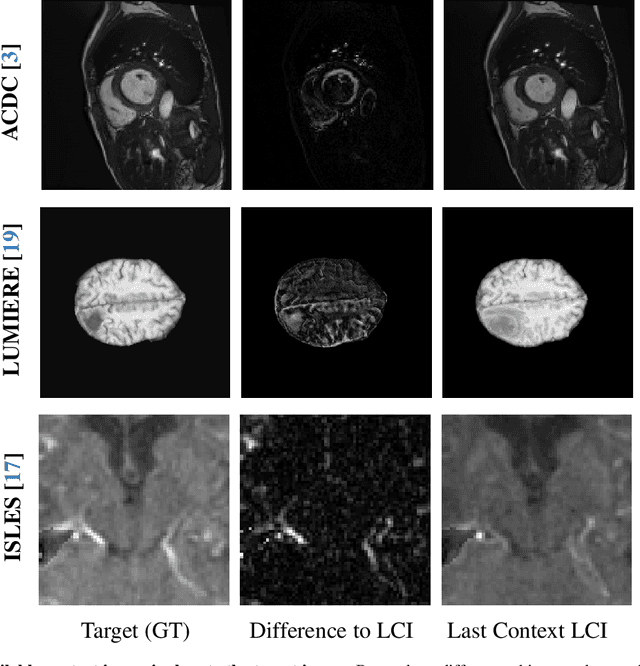
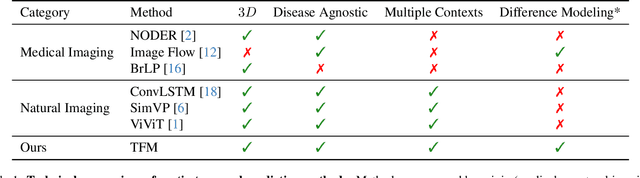
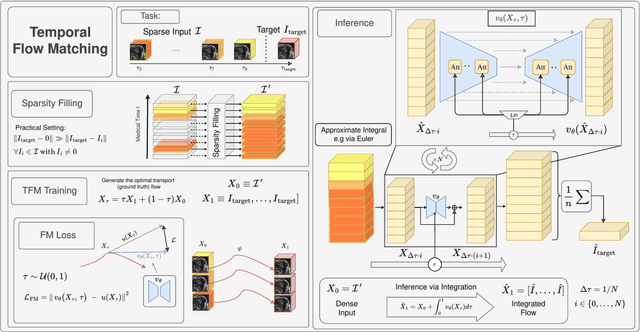
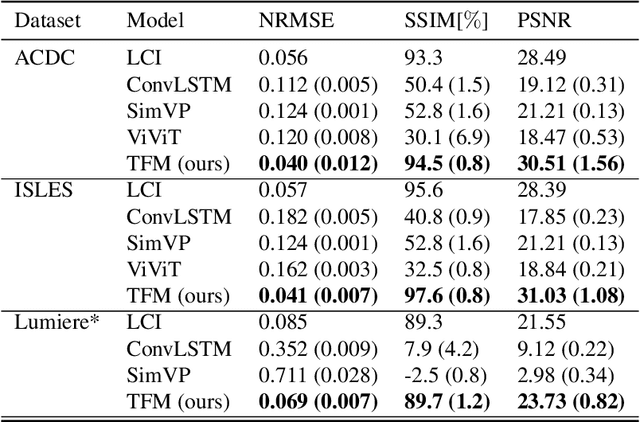
Abstract:Understanding temporal dynamics in medical imaging is crucial for applications such as disease progression modeling, treatment planning and anatomical development tracking. However, most deep learning methods either consider only single temporal contexts, or focus on tasks like classification or regression, limiting their ability for fine-grained spatial predictions. While some approaches have been explored, they are often limited to single timepoints, specific diseases or have other technical restrictions. To address this fundamental gap, we introduce Temporal Flow Matching (TFM), a unified generative trajectory method that (i) aims to learn the underlying temporal distribution, (ii) by design can fall back to a nearest image predictor, i.e. predicting the last context image (LCI), as a special case, and (iii) supports $3D$ volumes, multiple prior scans, and irregular sampling. Extensive benchmarks on three public longitudinal datasets show that TFM consistently surpasses spatio-temporal methods from natural imaging, establishing a new state-of-the-art and robust baseline for $4D$ medical image prediction.
nnInteractive: Redefining 3D Promptable Segmentation
Mar 11, 2025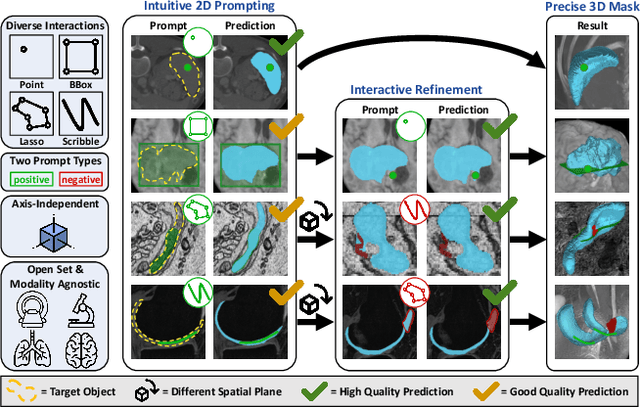
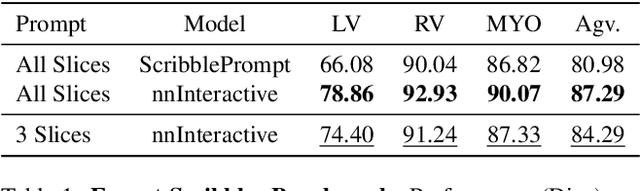
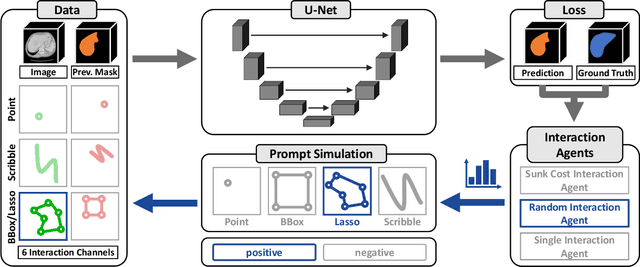
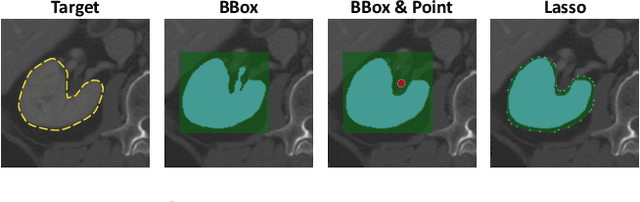
Abstract:Accurate and efficient 3D segmentation is essential for both clinical and research applications. While foundation models like SAM have revolutionized interactive segmentation, their 2D design and domain shift limitations make them ill-suited for 3D medical images. Current adaptations address some of these challenges but remain limited, either lacking volumetric awareness, offering restricted interactivity, or supporting only a small set of structures and modalities. Usability also remains a challenge, as current tools are rarely integrated into established imaging platforms and often rely on cumbersome web-based interfaces with restricted functionality. We introduce nnInteractive, the first comprehensive 3D interactive open-set segmentation method. It supports diverse prompts-including points, scribbles, boxes, and a novel lasso prompt-while leveraging intuitive 2D interactions to generate full 3D segmentations. Trained on 120+ diverse volumetric 3D datasets (CT, MRI, PET, 3D Microscopy, etc.), nnInteractive sets a new state-of-the-art in accuracy, adaptability, and usability. Crucially, it is the first method integrated into widely used image viewers (e.g., Napari, MITK), ensuring broad accessibility for real-world clinical and research applications. Extensive benchmarking demonstrates that nnInteractive far surpasses existing methods, setting a new standard for AI-driven interactive 3D segmentation. nnInteractive is publicly available: https://github.com/MIC-DKFZ/napari-nninteractive (Napari plugin), https://www.mitk.org/MITK-nnInteractive (MITK integration), https://github.com/MIC-DKFZ/nnInteractive (Python backend).
LesionLocator: Zero-Shot Universal Tumor Segmentation and Tracking in 3D Whole-Body Imaging
Feb 28, 2025Abstract:In this work, we present LesionLocator, a framework for zero-shot longitudinal lesion tracking and segmentation in 3D medical imaging, establishing the first end-to-end model capable of 4D tracking with dense spatial prompts. Our model leverages an extensive dataset of 23,262 annotated medical scans, as well as synthesized longitudinal data across diverse lesion types. The diversity and scale of our dataset significantly enhances model generalizability to real-world medical imaging challenges and addresses key limitations in longitudinal data availability. LesionLocator outperforms all existing promptable models in lesion segmentation by nearly 10 dice points, reaching human-level performance, and achieves state-of-the-art results in lesion tracking, with superior lesion retrieval and segmentation accuracy. LesionLocator not only sets a new benchmark in universal promptable lesion segmentation and automated longitudinal lesion tracking but also provides the first open-access solution of its kind, releasing our synthetic 4D dataset and model to the community, empowering future advancements in medical imaging. Code is available at: www.github.com/MIC-DKFZ/LesionLocator
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge