Heinz-Peter Schlemmer
LesionLocator: Zero-Shot Universal Tumor Segmentation and Tracking in 3D Whole-Body Imaging
Feb 28, 2025Abstract:In this work, we present LesionLocator, a framework for zero-shot longitudinal lesion tracking and segmentation in 3D medical imaging, establishing the first end-to-end model capable of 4D tracking with dense spatial prompts. Our model leverages an extensive dataset of 23,262 annotated medical scans, as well as synthesized longitudinal data across diverse lesion types. The diversity and scale of our dataset significantly enhances model generalizability to real-world medical imaging challenges and addresses key limitations in longitudinal data availability. LesionLocator outperforms all existing promptable models in lesion segmentation by nearly 10 dice points, reaching human-level performance, and achieves state-of-the-art results in lesion tracking, with superior lesion retrieval and segmentation accuracy. LesionLocator not only sets a new benchmark in universal promptable lesion segmentation and automated longitudinal lesion tracking but also provides the first open-access solution of its kind, releasing our synthetic 4D dataset and model to the community, empowering future advancements in medical imaging. Code is available at: www.github.com/MIC-DKFZ/LesionLocator
Pre-examinations Improve Automated Metastases Detection on Cranial MRI
Mar 13, 2024Abstract:Materials and methods: First, a dual-time approach was assessed, for which the CNN was provided sequences of the MRI that initially depicted new MM (diagnosis MRI) as well as of a prediagnosis MRI: inclusion of only contrast-enhanced T1-weighted images (CNNdual_ce) was compared with inclusion of also the native T1-weighted images, T2-weighted images, and FLAIR sequences of both time points (CNNdual_all).Second, results were compared with the corresponding single time approaches, in which the CNN was provided exclusively the respective sequences of the diagnosis MRI.Casewise diagnostic performance parameters were calculated from 5-fold cross-validation. Results: In total, 94 cases with 494 MMs were included. Overall, the highest diagnostic performance was achieved by inclusion of only the contrast-enhanced T1-weighted images of the diagnosis and of a prediagnosis MRI (CNNdual_ce, sensitivity = 73%, PPV = 25%, F1-score = 36%). Using exclusively contrast-enhanced T1-weighted images as input resulted in significantly less false-positives (FPs) compared with inclusion of further sequences beyond contrast-enhanced T1-weighted images (FPs = 5/7 for CNNdual_ce/CNNdual_all, P < 1e-5). Comparison of contrast-enhanced dual and mono time approaches revealed that exclusion of prediagnosis MRI significantly increased FPs (FPs = 5/10 for CNNdual_ce/CNNce, P < 1e-9).Approaches with only native sequences were clearly inferior to CNNs that were provided contrast-enhanced sequences. Conclusions: Automated MM detection on contrast-enhanced T1-weighted images performed with high sensitivity. Frequent FPs due to artifacts and vessels were significantly reduced by additional inclusion of prediagnosis MRI, but not by inclusion of further sequences beyond contrast-enhanced T1-weighted images. Future studies might investigate different change detection architectures for computer-aided detection.
Anatomy-informed Data Augmentation for Enhanced Prostate Cancer Detection
Sep 07, 2023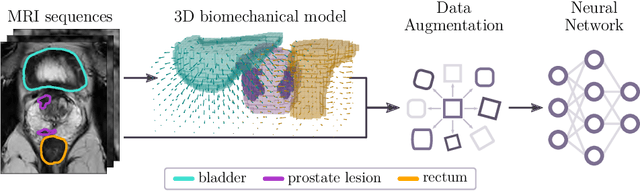

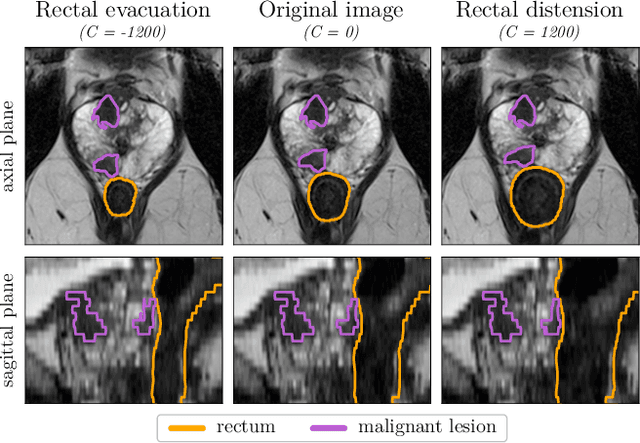
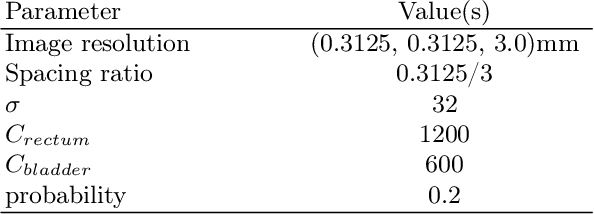
Abstract:Data augmentation (DA) is a key factor in medical image analysis, such as in prostate cancer (PCa) detection on magnetic resonance images. State-of-the-art computer-aided diagnosis systems still rely on simplistic spatial transformations to preserve the pathological label post transformation. However, such augmentations do not substantially increase the organ as well as tumor shape variability in the training set, limiting the model's ability to generalize to unseen cases with more diverse localized soft-tissue deformations. We propose a new anatomy-informed transformation that leverages information from adjacent organs to simulate typical physiological deformations of the prostate and generates unique lesion shapes without altering their label. Due to its lightweight computational requirements, it can be easily integrated into common DA frameworks. We demonstrate the effectiveness of our augmentation on a dataset of 774 biopsy-confirmed examinations, by evaluating a state-of-the-art method for PCa detection with different augmentation settings.
'A net for everyone': fully personalized and unsupervised neural networks trained with longitudinal data from a single patient
Oct 25, 2022Abstract:With the rise in importance of personalized medicine, we trained personalized neural networks to detect tumor progression in longitudinal datasets. The model was evaluated on two datasets with a total of 64 scans from 32 patients diagnosed with glioblastoma multiforme (GBM). Contrast-enhanced T1w sequences of brain magnetic resonance imaging (MRI) images were used in this study. For each patient, we trained their own neural network using just two images from different timepoints. Our approach uses a Wasserstein-GAN (generative adversarial network), an unsupervised network architecture, to map the differences between the two images. Using this map, the change in tumor volume can be evaluated. Due to the combination of data augmentation and the network architecture, co-registration of the two images is not needed. Furthermore, we do not rely on any additional training data, (manual) annotations or pre-training neural networks. The model received an AUC-score of 0.87 for tumor change. We also introduced a modified RANO criteria, for which an accuracy of 66% can be achieved. We show that using data from just one patient can be used to train deep neural networks to monitor tumor change.
Prediction of low-keV monochromatic images from polyenergetic CT scans for improved automatic detection of pulmonary embolism
Feb 23, 2021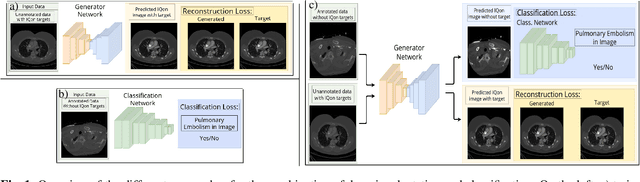
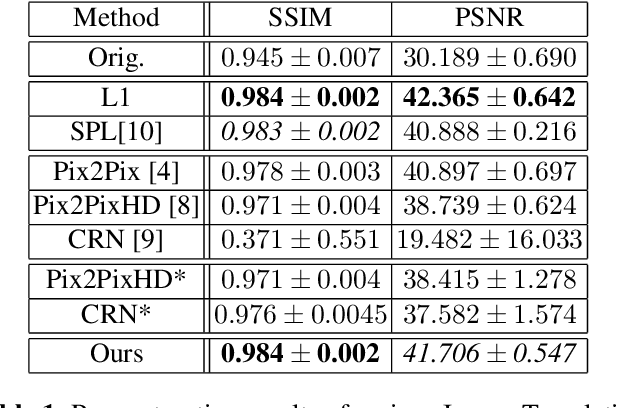
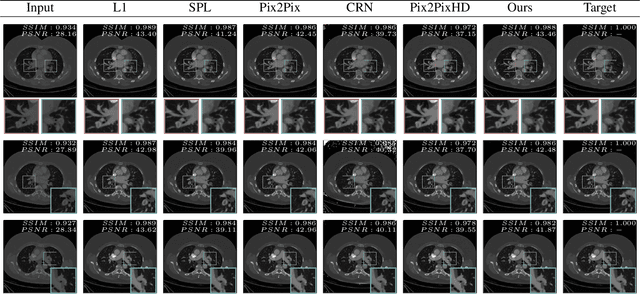
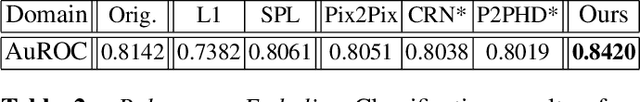
Abstract:Detector-based spectral computed tomography is a recent dual-energy CT (DECT) technology that offers the possibility of obtaining spectral information. From this spectral data, different types of images can be derived, amongst others virtual monoenergetic (monoE) images. MonoE images potentially exhibit decreased artifacts, improve contrast, and overall contain lower noise values, making them ideal candidates for better delineation and thus improved diagnostic accuracy of vascular abnormalities. In this paper, we are training convolutional neural networks~(CNN) that can emulate the generation of monoE images from conventional single energy CT acquisitions. For this task, we investigate several commonly used image-translation methods. We demonstrate that these methods while creating visually similar outputs, lead to a poorer performance when used for automatic classification of pulmonary embolism (PE). We expand on these methods through the use of a multi-task optimization approach, under which the networks achieve improved classification as well as generation results, as reflected by PSNR and SSIM scores. Further, evaluating our proposed framework on a subset of the RSNA-PE challenge data set shows that we are able to improve the Area under the Receiver Operating Characteristic curve (AuROC) in comparison to a na\"ive classification approach from 0.8142 to 0.8420.
Self-Guided Multiple Instance Learning for Weakly Supervised Disease Classification and Localization in Chest Radiographs
Sep 30, 2020



Abstract:The lack of fine-grained annotations hinders the deployment of automated diagnosis systems, which require human-interpretable justification for their decision process. In this paper, we address the problem of weakly supervised identification and localization of abnormalities in chest radiographs. To that end, we introduce a novel loss function for training convolutional neural networks increasing the \emph{localization confidence} and assisting the overall \emph{disease identification}. The loss leverages both image- and patch-level predictions to generate auxiliary supervision. Rather than forming strictly binary from the predictions as done in previous loss formulations, we create targets in a more customized manner, which allows the loss to account for possible misclassification. We show that the supervision provided within the proposed learning scheme leads to better performance and more precise predictions on prevalent datasets for multiple-instance learning as well as on the NIH~ChestX-Ray14 benchmark for disease recognition than previously used losses.
Automated brain extraction of multi-sequence MRI using artificial neural networks
Jan 31, 2019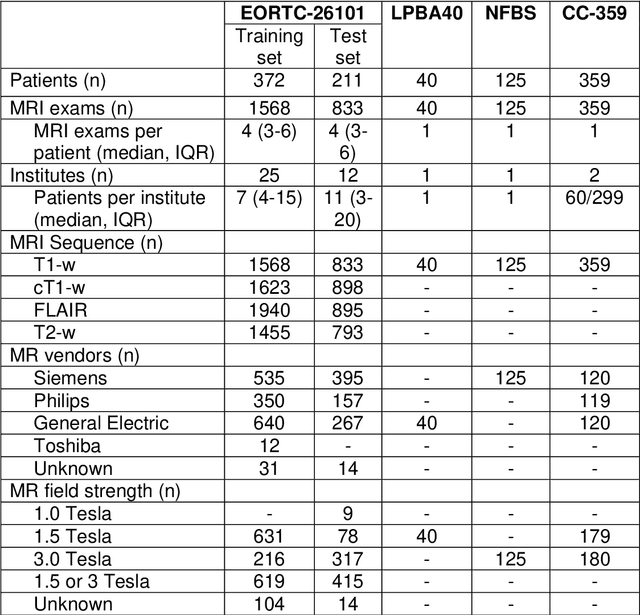
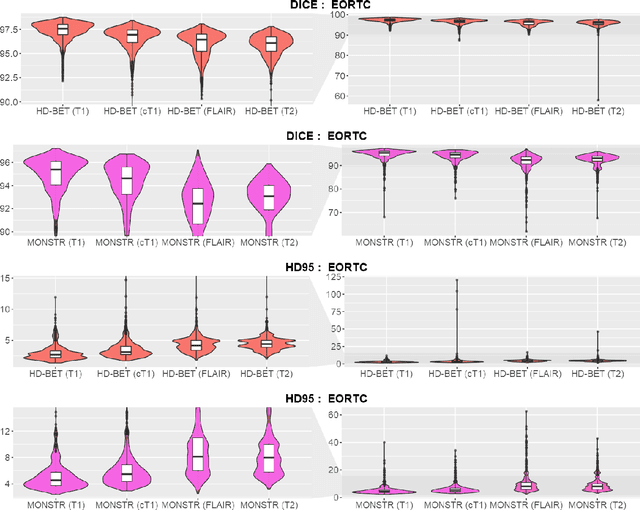
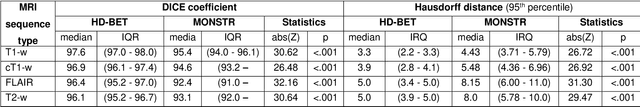
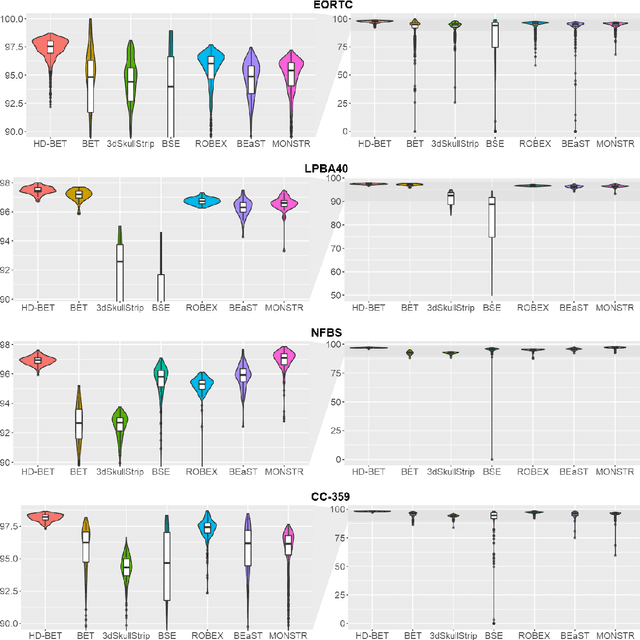
Abstract:Brain extraction is a critical preprocessing step in the analysis of MRI neuroimaging studies and influences the accuracy of downstream analyses. State-of-the-art brain extraction algorithms are, however, optimized for processing healthy brains and thus frequently fail in the presence of pathologically altered brain or when applied to heterogeneous MRI datasets. Here we introduce a new, rigorously validated algorithm (termed HD-BET) relying on artificial neural networks that aims to overcome these limitations. We demonstrate that HD-BET outperforms five publicly available state-of-the-art brain extraction algorithms in several large-scale neuroimaging datasets, including one from a prospective multicentric trial in neuro-oncology, yielding median improvements of +1.33 to +2.63 points for the DICE coefficient and -0.80 to -2.75 mm for the Hausdorff distance (Bonferroni-adjusted p<0.001). Importantly, the HD-BET algorithm shows robust performance in the presence of pathology or treatment-induced tissue alterations, is applicable to a broad range of MRI sequence types and is not influenced by variations in MRI hardware and acquisition parameters encountered in both research and clinical practice. For broader accessibility our HD-BET prediction algorithm is made freely available and may become an essential component for robust, automated, high-throughput processing of MRI neuroimaging data.
Retina U-Net: Embarrassingly Simple Exploitation of Segmentation Supervision for Medical Object Detection
Nov 21, 2018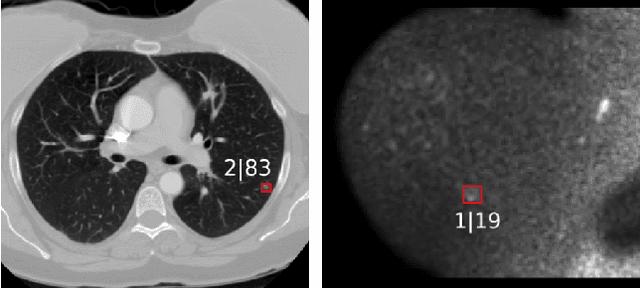
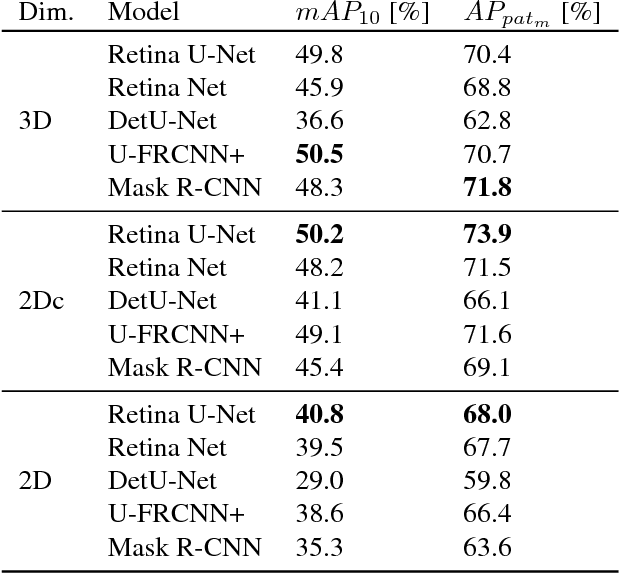
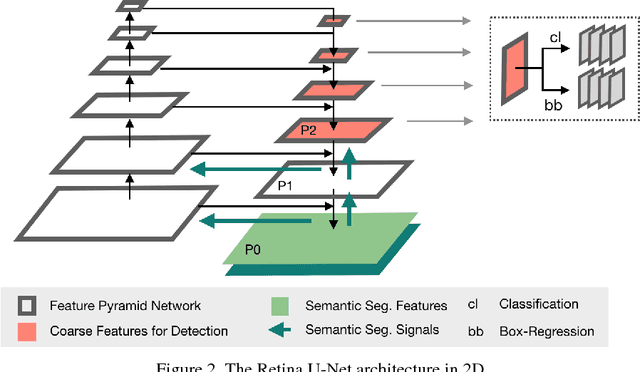
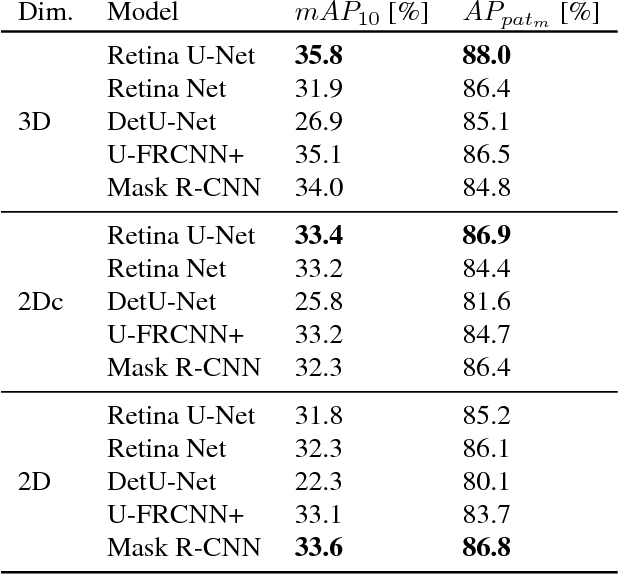
Abstract:The task of localizing and categorizing objects in medical images often remains formulated as a semantic segmentation problem. This approach, however, only indirectly solves the coarse localization task by predicting pixel-level scores, requiring ad-hoc heuristics when mapping back to object-level scores. State-of-the-art object detectors on the other hand, allow for individual object scoring in an end-to-end fashion, while ironically trading in the ability to exploit the full pixel-wise supervision signal. This can be particularly disadvantageous in the setting of medical image analysis, where data sets are notoriously small. In this paper, we propose Retina U-Net, a simple architecture, which naturally fuses the Retina Net one-stage detector with the U-Net architecture widely used for semantic segmentation in medical images. The proposed architecture recaptures discarded supervision signals by complementing object detection with an auxiliary task in the form of semantic segmentation without introducing the additional complexity of previously proposed two-stage detectors. We evaluate the importance of full segmentation supervision on two medical data sets, provide an in-depth analysis on a series of toy experiments and show how the corresponding performance gain grows in the limit of small data sets. Retina U-Net yields strong detection performance only reached by its more complex two-staged counterparts. Our framework including all methods implemented for operation on 2D and 3D images is available at github.com/pfjaeger/medicaldetectiontoolkit.
Domain Adaptation for Deviating Acquisition Protocols in CNN-based Lesion Classification on Diffusion-Weighted MR Images
Jul 17, 2018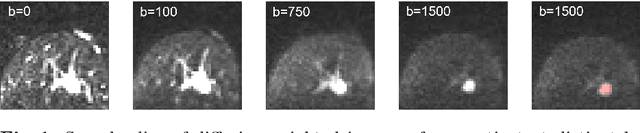
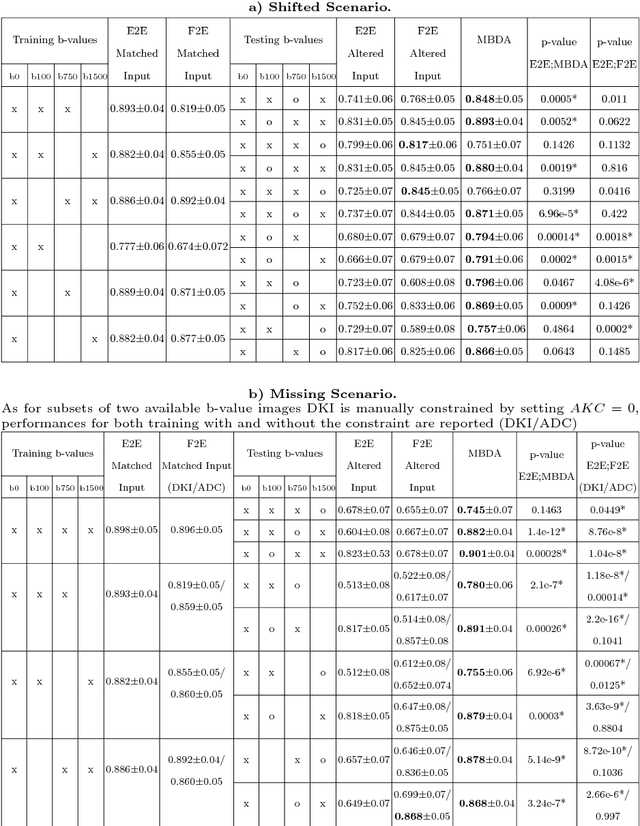
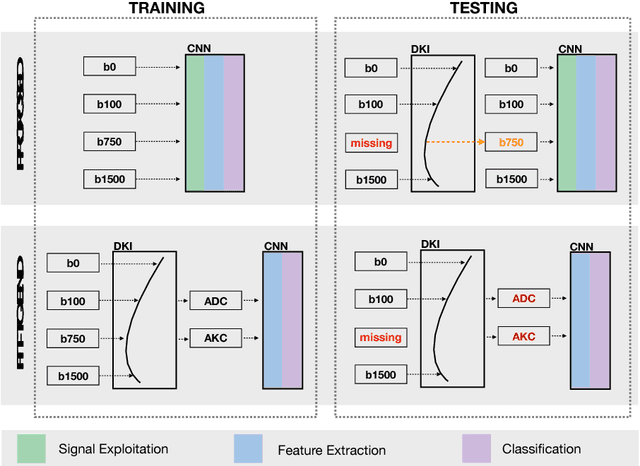
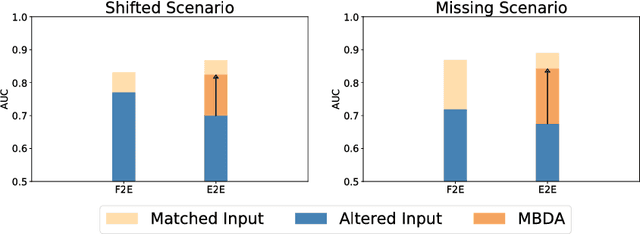
Abstract:End-to-end deep learning improves breast cancer classification on diffusion-weighted MR images (DWI) using a convolutional neural network (CNN) architecture. A limitation of CNN as opposed to previous model-based approaches is the dependence on specific DWI input channels used during training. However, in the context of large-scale application, methods agnostic towards heterogeneous inputs are desirable, due to the high deviation of scanning protocols between clinical sites. We propose model-based domain adaptation to overcome input dependencies and avoid re-training of networks at clinical sites by restoring training inputs from altered input channels given during deployment. We demonstrate the method's significant increase in classification performance and superiority over implicit domain adaptation provided by training-schemes operating on model-parameters instead of raw DWI images.
Adversarial Networks for Prostate Cancer Detection
Nov 28, 2017


Abstract:The large number of trainable parameters of deep neural networks renders them inherently data hungry. This characteristic heavily challenges the medical imaging community and to make things even worse, many imaging modalities are ambiguous in nature leading to rater-dependant annotations that current loss formulations fail to capture. We propose employing adversarial training for segmentation networks in order to alleviate aforementioned problems. We learn to segment aggressive prostate cancer utilizing challenging MRI images of 152 patients and show that the proposed scheme is superior over the de facto standard in terms of the detection sensitivity and the dice-score for aggressive prostate cancer. The achieved relative gains are shown to be particularly pronounced in the small dataset limit.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge