Ulf Neuberger
Continuous-Time Deep Glioma Growth Models
Jul 02, 2021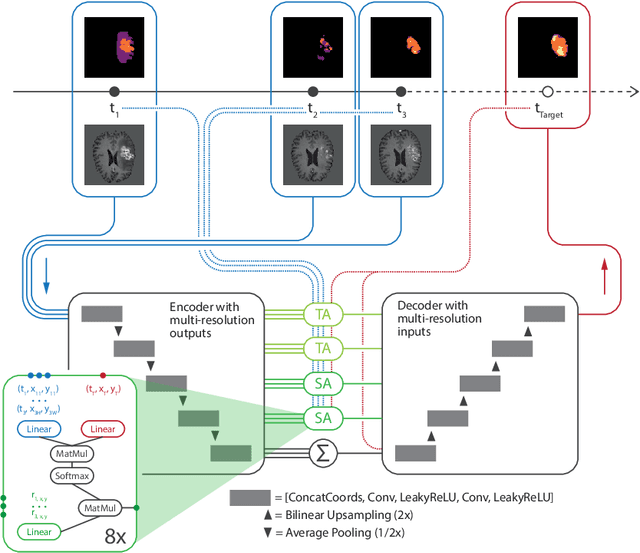
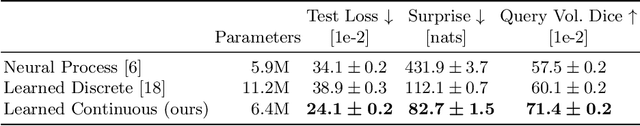

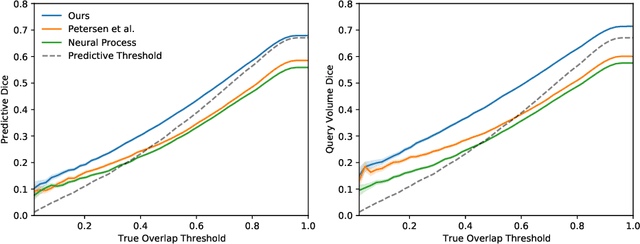
Abstract:The ability to estimate how a tumor might evolve in the future could have tremendous clinical benefits, from improved treatment decisions to better dose distribution in radiation therapy. Recent work has approached the glioma growth modeling problem via deep learning and variational inference, thus learning growth dynamics entirely from a real patient data distribution. So far, this approach was constrained to predefined image acquisition intervals and sequences of fixed length, which limits its applicability in more realistic scenarios. We overcome these limitations by extending Neural Processes, a class of conditional generative models for stochastic time series, with a hierarchical multi-scale representation encoding including a spatio-temporal attention mechanism. The result is a learned growth model that can be conditioned on an arbitrary number of observations, and that can produce a distribution of temporally consistent growth trajectories on a continuous time axis. On a dataset of 379 patients, the approach successfully captures both global and finer-grained variations in the images, exhibiting superior performance compared to other learned growth models.
Deep Probabilistic Modeling of Glioma Growth
Jul 09, 2019


Abstract:Existing approaches to modeling the dynamics of brain tumor growth, specifically glioma, employ biologically inspired models of cell diffusion, using image data to estimate the associated parameters. In this work, we propose an alternative approach based on recent advances in probabilistic segmentation and representation learning that implicitly learns growth dynamics directly from data without an underlying explicit model. We present evidence that our approach is able to learn a distribution of plausible future tumor appearances conditioned on past observations of the same tumor.
Automated brain extraction of multi-sequence MRI using artificial neural networks
Jan 31, 2019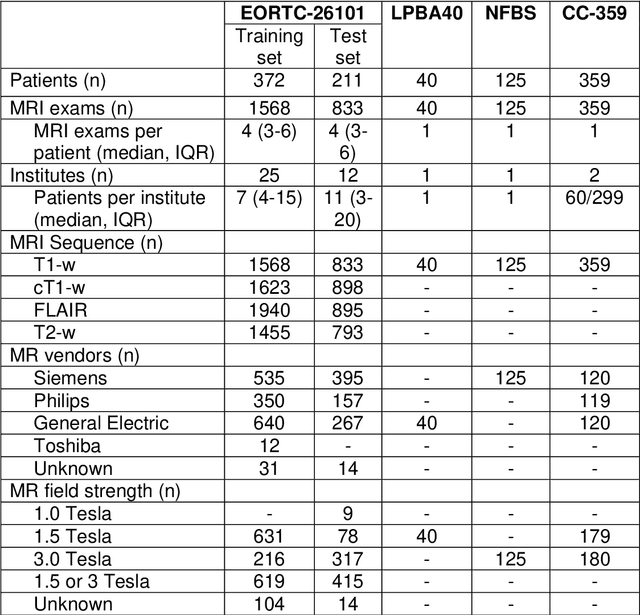
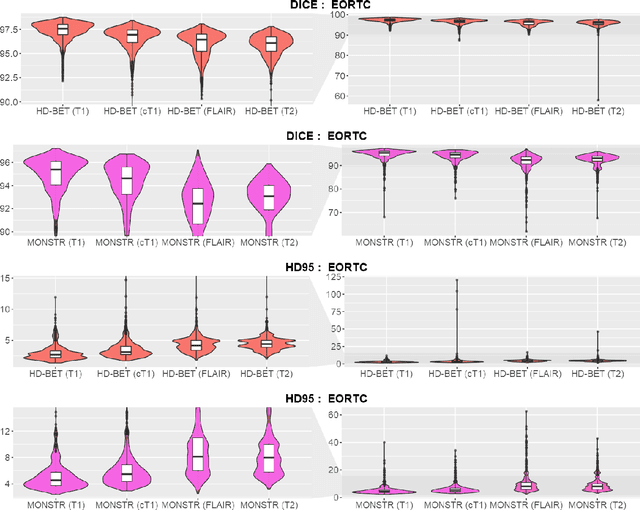
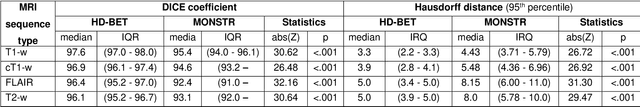
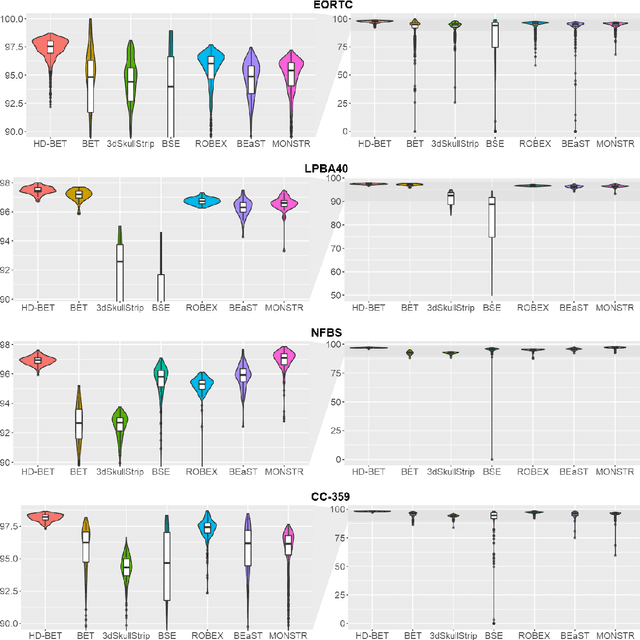
Abstract:Brain extraction is a critical preprocessing step in the analysis of MRI neuroimaging studies and influences the accuracy of downstream analyses. State-of-the-art brain extraction algorithms are, however, optimized for processing healthy brains and thus frequently fail in the presence of pathologically altered brain or when applied to heterogeneous MRI datasets. Here we introduce a new, rigorously validated algorithm (termed HD-BET) relying on artificial neural networks that aims to overcome these limitations. We demonstrate that HD-BET outperforms five publicly available state-of-the-art brain extraction algorithms in several large-scale neuroimaging datasets, including one from a prospective multicentric trial in neuro-oncology, yielding median improvements of +1.33 to +2.63 points for the DICE coefficient and -0.80 to -2.75 mm for the Hausdorff distance (Bonferroni-adjusted p<0.001). Importantly, the HD-BET algorithm shows robust performance in the presence of pathology or treatment-induced tissue alterations, is applicable to a broad range of MRI sequence types and is not influenced by variations in MRI hardware and acquisition parameters encountered in both research and clinical practice. For broader accessibility our HD-BET prediction algorithm is made freely available and may become an essential component for robust, automated, high-throughput processing of MRI neuroimaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge