David Bonekamp
Anatomy-informed Data Augmentation for Enhanced Prostate Cancer Detection
Sep 07, 2023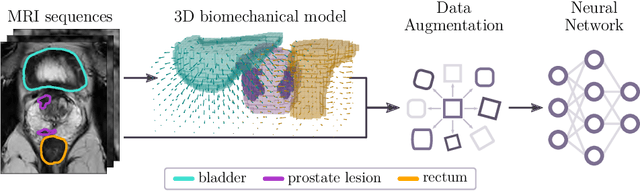

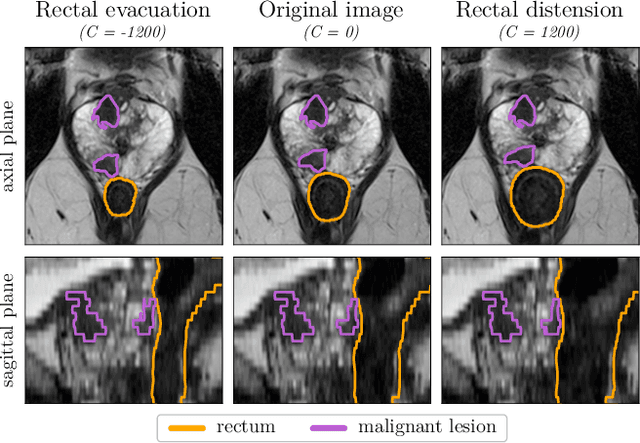
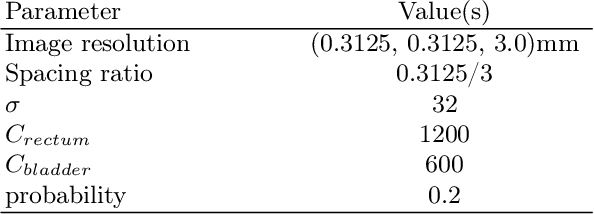
Abstract:Data augmentation (DA) is a key factor in medical image analysis, such as in prostate cancer (PCa) detection on magnetic resonance images. State-of-the-art computer-aided diagnosis systems still rely on simplistic spatial transformations to preserve the pathological label post transformation. However, such augmentations do not substantially increase the organ as well as tumor shape variability in the training set, limiting the model's ability to generalize to unseen cases with more diverse localized soft-tissue deformations. We propose a new anatomy-informed transformation that leverages information from adjacent organs to simulate typical physiological deformations of the prostate and generates unique lesion shapes without altering their label. Due to its lightweight computational requirements, it can be easily integrated into common DA frameworks. We demonstrate the effectiveness of our augmentation on a dataset of 774 biopsy-confirmed examinations, by evaluating a state-of-the-art method for PCa detection with different augmentation settings.
Automated brain extraction of multi-sequence MRI using artificial neural networks
Jan 31, 2019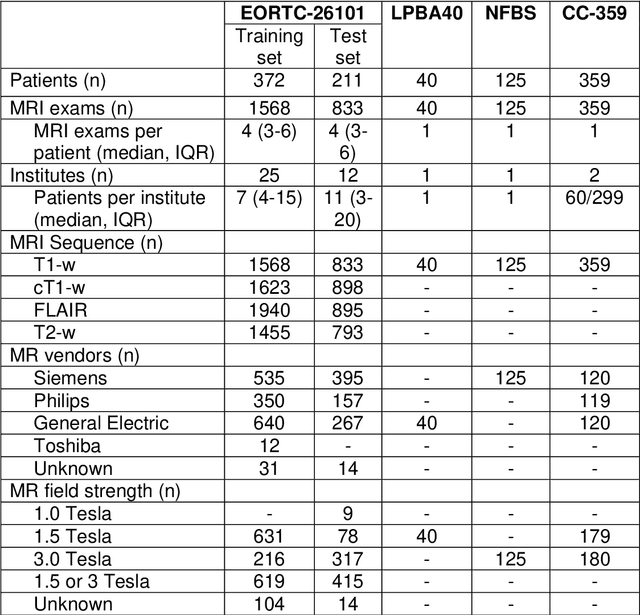
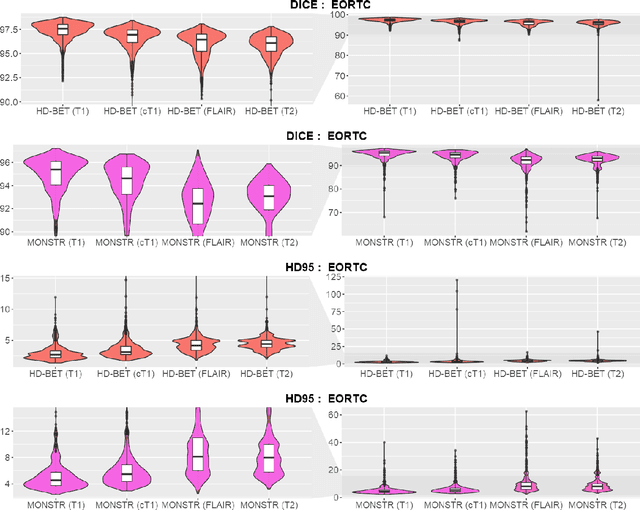
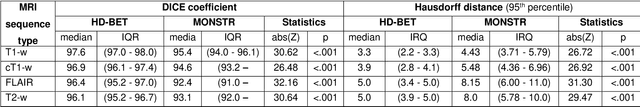
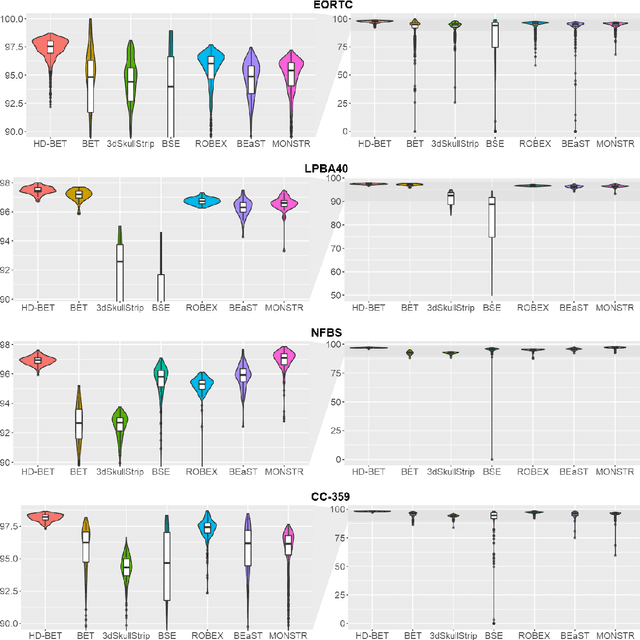
Abstract:Brain extraction is a critical preprocessing step in the analysis of MRI neuroimaging studies and influences the accuracy of downstream analyses. State-of-the-art brain extraction algorithms are, however, optimized for processing healthy brains and thus frequently fail in the presence of pathologically altered brain or when applied to heterogeneous MRI datasets. Here we introduce a new, rigorously validated algorithm (termed HD-BET) relying on artificial neural networks that aims to overcome these limitations. We demonstrate that HD-BET outperforms five publicly available state-of-the-art brain extraction algorithms in several large-scale neuroimaging datasets, including one from a prospective multicentric trial in neuro-oncology, yielding median improvements of +1.33 to +2.63 points for the DICE coefficient and -0.80 to -2.75 mm for the Hausdorff distance (Bonferroni-adjusted p<0.001). Importantly, the HD-BET algorithm shows robust performance in the presence of pathology or treatment-induced tissue alterations, is applicable to a broad range of MRI sequence types and is not influenced by variations in MRI hardware and acquisition parameters encountered in both research and clinical practice. For broader accessibility our HD-BET prediction algorithm is made freely available and may become an essential component for robust, automated, high-throughput processing of MRI neuroimaging data.
Adversarial Networks for Prostate Cancer Detection
Nov 28, 2017


Abstract:The large number of trainable parameters of deep neural networks renders them inherently data hungry. This characteristic heavily challenges the medical imaging community and to make things even worse, many imaging modalities are ambiguous in nature leading to rater-dependant annotations that current loss formulations fail to capture. We propose employing adversarial training for segmentation networks in order to alleviate aforementioned problems. We learn to segment aggressive prostate cancer utilizing challenging MRI images of 152 patients and show that the proposed scheme is superior over the de facto standard in terms of the detection sensitivity and the dice-score for aggressive prostate cancer. The achieved relative gains are shown to be particularly pronounced in the small dataset limit.
Revealing Hidden Potentials of the q-Space Signal in Breast Cancer
May 22, 2017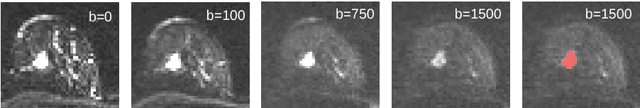

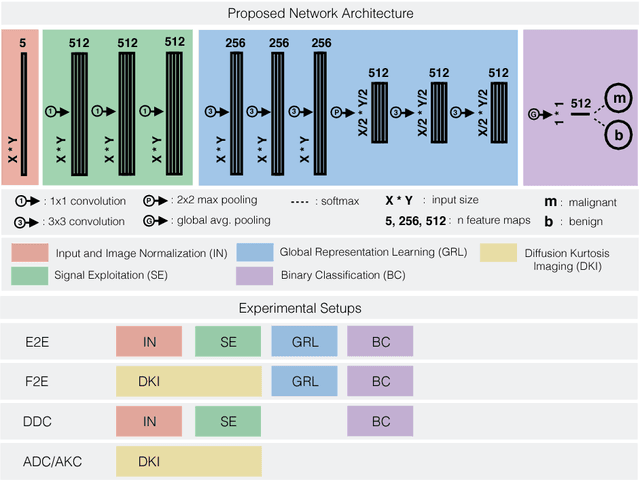
Abstract:Mammography screening for early detection of breast lesions currently suffers from high amounts of false positive findings, which result in unnecessary invasive biopsies. Diffusion-weighted MR images (DWI) can help to reduce many of these false-positive findings prior to biopsy. Current approaches estimate tissue properties by means of quantitative parameters taken from generative, biophysical models fit to the q-space encoded signal under certain assumptions regarding noise and spatial homogeneity. This process is prone to fitting instability and partial information loss due to model simplicity. We reveal unexplored potentials of the signal by integrating all data processing components into a convolutional neural network (CNN) architecture that is designed to propagate clinical target information down to the raw input images. This approach enables simultaneous and target-specific optimization of image normalization, signal exploitation, global representation learning and classification. Using a multicentric data set of 222 patients, we demonstrate that our approach significantly improves clinical decision making with respect to the current state of the art.
Adversarial Networks for the Detection of Aggressive Prostate Cancer
Feb 26, 2017



Abstract:Semantic segmentation constitutes an integral part of medical image analyses for which breakthroughs in the field of deep learning were of high relevance. The large number of trainable parameters of deep neural networks however renders them inherently data hungry, a characteristic that heavily challenges the medical imaging community. Though interestingly, with the de facto standard training of fully convolutional networks (FCNs) for semantic segmentation being agnostic towards the `structure' of the predicted label maps, valuable complementary information about the global quality of the segmentation lies idle. In order to tap into this potential, we propose utilizing an adversarial network which discriminates between expert and generated annotations in order to train FCNs for semantic segmentation. Because the adversary constitutes a learned parametrization of what makes a good segmentation at a global level, we hypothesize that the method holds particular advantages for segmentation tasks on complex structured, small datasets. This holds true in our experiments: We learn to segment aggressive prostate cancer utilizing MRI images of 152 patients and show that the proposed scheme is superior over the de facto standard in terms of the detection sensitivity and the dice-score for aggressive prostate cancer. The achieved relative gains are shown to be particularly pronounced in the small dataset limit.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge