Balint Kovacs
LesionLocator: Zero-Shot Universal Tumor Segmentation and Tracking in 3D Whole-Body Imaging
Feb 28, 2025Abstract:In this work, we present LesionLocator, a framework for zero-shot longitudinal lesion tracking and segmentation in 3D medical imaging, establishing the first end-to-end model capable of 4D tracking with dense spatial prompts. Our model leverages an extensive dataset of 23,262 annotated medical scans, as well as synthesized longitudinal data across diverse lesion types. The diversity and scale of our dataset significantly enhances model generalizability to real-world medical imaging challenges and addresses key limitations in longitudinal data availability. LesionLocator outperforms all existing promptable models in lesion segmentation by nearly 10 dice points, reaching human-level performance, and achieves state-of-the-art results in lesion tracking, with superior lesion retrieval and segmentation accuracy. LesionLocator not only sets a new benchmark in universal promptable lesion segmentation and automated longitudinal lesion tracking but also provides the first open-access solution of its kind, releasing our synthetic 4D dataset and model to the community, empowering future advancements in medical imaging. Code is available at: www.github.com/MIC-DKFZ/LesionLocator
Data-Centric Strategies for Overcoming PET/CT Heterogeneity: Insights from the AutoPET III Lesion Segmentation Challenge
Sep 16, 2024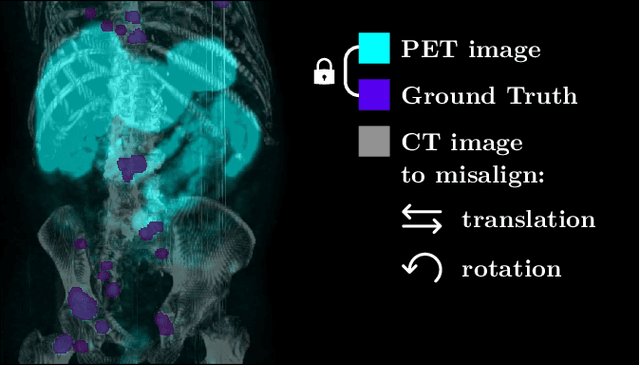

Abstract:The third autoPET challenge introduced a new data-centric task this year, shifting the focus from model development to improving metastatic lesion segmentation on PET/CT images through data quality and handling strategies. In response, we developed targeted methods to enhance segmentation performance tailored to the characteristics of PET/CT imaging. Our approach encompasses two key elements. First, to address potential alignment errors between CT and PET modalities as well as the prevalence of punctate lesions, we modified the baseline data augmentation scheme and extended it with misalignment augmentation. This adaptation aims to improve segmentation accuracy, particularly for tiny metastatic lesions. Second, to tackle the variability in image dimensions significantly affecting the prediction time, we implemented a dynamic ensembling and test-time augmentation (TTA) strategy. This method optimizes the use of ensembling and TTA within a 5-minute prediction time limit, effectively leveraging the generalization potential for both small and large images. Both of our solutions are designed to be robust across different tracers and institutional settings, offering a general, yet imaging-specific approach to the multi-tracer and multi-institutional challenges of the competition. We made the challenge repository with our modifications publicly available at \url{https://github.com/MIC-DKFZ/miccai2024_autopet3_datacentric}.
From FDG to PSMA: A Hitchhiker's Guide to Multitracer, Multicenter Lesion Segmentation in PET/CT Imaging
Sep 14, 2024

Abstract:Automated lesion segmentation in PET/CT scans is crucial for improving clinical workflows and advancing cancer diagnostics. However, the task is challenging due to physiological variability, different tracers used in PET imaging, and diverse imaging protocols across medical centers. To address this, the autoPET series was created to challenge researchers to develop algorithms that generalize across diverse PET/CT environments. This paper presents our solution for the autoPET III challenge, targeting multitracer, multicenter generalization using the nnU-Net framework with the ResEncL architecture. Key techniques include misalignment data augmentation and multi-modal pretraining across CT, MR, and PET datasets to provide an initial anatomical understanding. We incorporate organ supervision as a multitask approach, enabling the model to distinguish between physiological uptake and tracer-specific patterns, which is particularly beneficial in cases where no lesions are present. Compared to the default nnU-Net, which achieved a Dice score of 57.61, or the larger ResEncL (65.31) our model significantly improved performance with a Dice score of 68.40, alongside a reduction in false positive (FPvol: 7.82) and false negative (FNvol: 10.35) volumes. These results underscore the effectiveness of combining advanced network design, augmentation, pretraining, and multitask learning for PET/CT lesion segmentation. Code is publicly available at https://github.com/MIC-DKFZ/autopet-3-submission.
Skeleton Recall Loss for Connectivity Conserving and Resource Efficient Segmentation of Thin Tubular Structures
Apr 03, 2024



Abstract:Accurately segmenting thin tubular structures, such as vessels, nerves, roads or concrete cracks, is a crucial task in computer vision. Standard deep learning-based segmentation loss functions, such as Dice or Cross-Entropy, focus on volumetric overlap, often at the expense of preserving structural connectivity or topology. This can lead to segmentation errors that adversely affect downstream tasks, including flow calculation, navigation, and structural inspection. Although current topology-focused losses mark an improvement, they introduce significant computational and memory overheads. This is particularly relevant for 3D data, rendering these losses infeasible for larger volumes as well as increasingly important multi-class segmentation problems. To mitigate this, we propose a novel Skeleton Recall Loss, which effectively addresses these challenges by circumventing intensive GPU-based calculations with inexpensive CPU operations. It demonstrates overall superior performance to current state-of-the-art approaches on five public datasets for topology-preserving segmentation, while substantially reducing computational overheads by more than 90%. In doing so, we introduce the first multi-class capable loss function for thin structure segmentation, excelling in both efficiency and efficacy for topology-preservation.
Anatomy-informed Data Augmentation for Enhanced Prostate Cancer Detection
Sep 07, 2023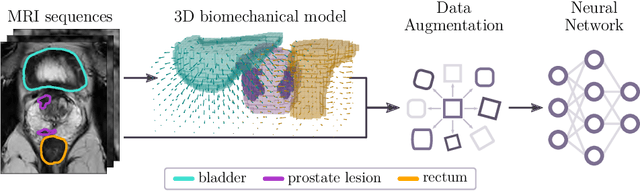

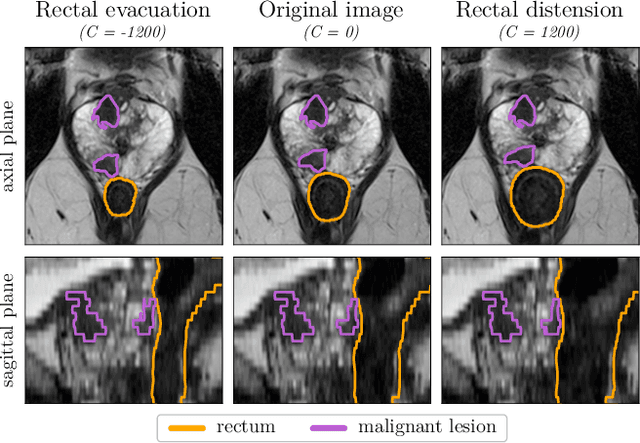
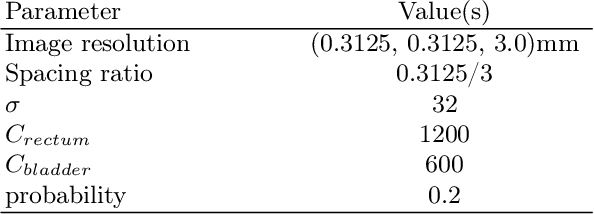
Abstract:Data augmentation (DA) is a key factor in medical image analysis, such as in prostate cancer (PCa) detection on magnetic resonance images. State-of-the-art computer-aided diagnosis systems still rely on simplistic spatial transformations to preserve the pathological label post transformation. However, such augmentations do not substantially increase the organ as well as tumor shape variability in the training set, limiting the model's ability to generalize to unseen cases with more diverse localized soft-tissue deformations. We propose a new anatomy-informed transformation that leverages information from adjacent organs to simulate typical physiological deformations of the prostate and generates unique lesion shapes without altering their label. Due to its lightweight computational requirements, it can be easily integrated into common DA frameworks. We demonstrate the effectiveness of our augmentation on a dataset of 774 biopsy-confirmed examinations, by evaluating a state-of-the-art method for PCa detection with different augmentation settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge