Maximilian R. Rokuss
Mitigating False Predictions In Unreasonable Body Regions
Apr 24, 2024
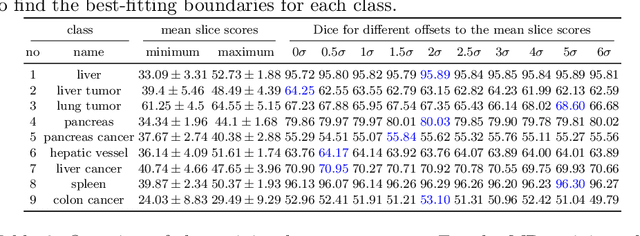


Abstract:Despite considerable strides in developing deep learning models for 3D medical image segmentation, the challenge of effectively generalizing across diverse image distributions persists. While domain generalization is acknowledged as vital for robust application in clinical settings, the challenges stemming from training with a limited Field of View (FOV) remain unaddressed. This limitation leads to false predictions when applied to body regions beyond the FOV of the training data. In response to this problem, we propose a novel loss function that penalizes predictions in implausible body regions, applicable in both single-dataset and multi-dataset training schemes. It is realized with a Body Part Regression model that generates axial slice positional scores. Through comprehensive evaluation using a test set featuring varying FOVs, our approach demonstrates remarkable improvements in generalization capabilities. It effectively mitigates false positive tumor predictions up to 85% and significantly enhances overall segmentation performance.
Skeleton Recall Loss for Connectivity Conserving and Resource Efficient Segmentation of Thin Tubular Structures
Apr 03, 2024



Abstract:Accurately segmenting thin tubular structures, such as vessels, nerves, roads or concrete cracks, is a crucial task in computer vision. Standard deep learning-based segmentation loss functions, such as Dice or Cross-Entropy, focus on volumetric overlap, often at the expense of preserving structural connectivity or topology. This can lead to segmentation errors that adversely affect downstream tasks, including flow calculation, navigation, and structural inspection. Although current topology-focused losses mark an improvement, they introduce significant computational and memory overheads. This is particularly relevant for 3D data, rendering these losses infeasible for larger volumes as well as increasingly important multi-class segmentation problems. To mitigate this, we propose a novel Skeleton Recall Loss, which effectively addresses these challenges by circumventing intensive GPU-based calculations with inexpensive CPU operations. It demonstrates overall superior performance to current state-of-the-art approaches on five public datasets for topology-preserving segmentation, while substantially reducing computational overheads by more than 90%. In doing so, we introduce the first multi-class capable loss function for thin structure segmentation, excelling in both efficiency and efficacy for topology-preservation.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
SAM.MD: Zero-shot medical image segmentation capabilities of the Segment Anything Model
Apr 10, 2023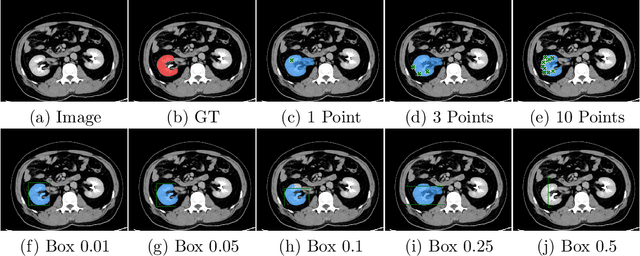
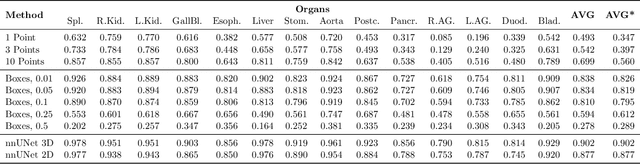
Abstract:Foundation models have taken over natural language processing and image generation domains due to the flexibility of prompting. With the recent introduction of the Segment Anything Model (SAM), this prompt-driven paradigm has entered image segmentation with a hitherto unexplored abundance of capabilities. The purpose of this paper is to conduct an initial evaluation of the out-of-the-box zero-shot capabilities of SAM for medical image segmentation, by evaluating its performance on an abdominal CT organ segmentation task, via point or bounding box based prompting. We show that SAM generalizes well to CT data, making it a potential catalyst for the advancement of semi-automatic segmentation tools for clinicians. We believe that this foundation model, while not reaching state-of-the-art segmentation performance in our investigations, can serve as a highly potent starting point for further adaptations of such models to the intricacies of the medical domain. Keywords: medical image segmentation, SAM, foundation models, zero-shot learning
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge