Martin Bendszus
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
The 2024 Brain Tumor Segmentation (BraTS) Challenge: Glioma Segmentation on Post-treatment MRI
May 28, 2024
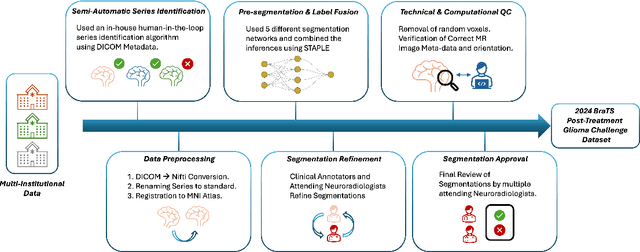
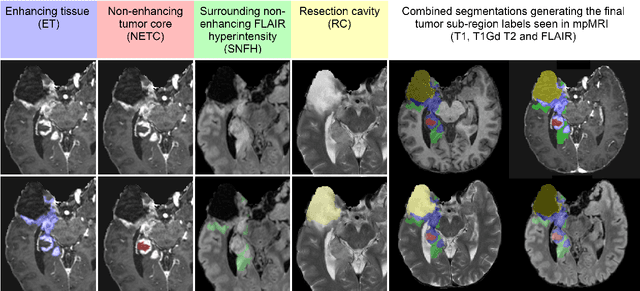
Abstract:Gliomas are the most common malignant primary brain tumors in adults and one of the deadliest types of cancer. There are many challenges in treatment and monitoring due to the genetic diversity and high intrinsic heterogeneity in appearance, shape, histology, and treatment response. Treatments include surgery, radiation, and systemic therapies, with magnetic resonance imaging (MRI) playing a key role in treatment planning and post-treatment longitudinal assessment. The 2024 Brain Tumor Segmentation (BraTS) challenge on post-treatment glioma MRI will provide a community standard and benchmark for state-of-the-art automated segmentation models based on the largest expert-annotated post-treatment glioma MRI dataset. Challenge competitors will develop automated segmentation models to predict four distinct tumor sub-regions consisting of enhancing tissue (ET), surrounding non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) hyperintensity (SNFH), non-enhancing tumor core (NETC), and resection cavity (RC). Models will be evaluated on separate validation and test datasets using standardized performance metrics utilized across the BraTS 2024 cluster of challenges, including lesion-wise Dice Similarity Coefficient and Hausdorff Distance. Models developed during this challenge will advance the field of automated MRI segmentation and contribute to their integration into clinical practice, ultimately enhancing patient care.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Continuous-Time Deep Glioma Growth Models
Jul 02, 2021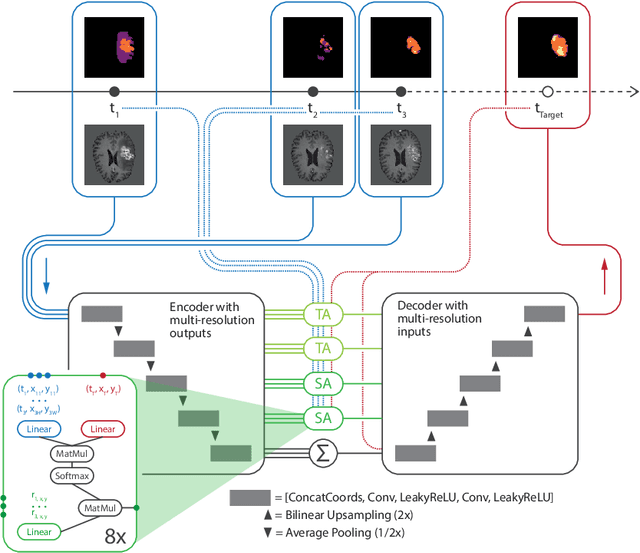
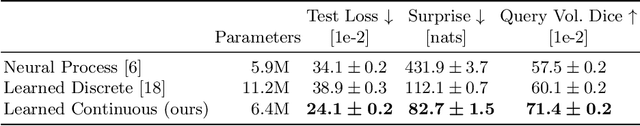

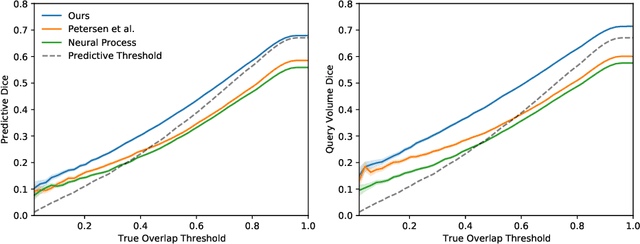
Abstract:The ability to estimate how a tumor might evolve in the future could have tremendous clinical benefits, from improved treatment decisions to better dose distribution in radiation therapy. Recent work has approached the glioma growth modeling problem via deep learning and variational inference, thus learning growth dynamics entirely from a real patient data distribution. So far, this approach was constrained to predefined image acquisition intervals and sequences of fixed length, which limits its applicability in more realistic scenarios. We overcome these limitations by extending Neural Processes, a class of conditional generative models for stochastic time series, with a hierarchical multi-scale representation encoding including a spatio-temporal attention mechanism. The result is a learned growth model that can be conditioned on an arbitrary number of observations, and that can produce a distribution of temporally consistent growth trajectories on a continuous time axis. On a dataset of 379 patients, the approach successfully captures both global and finer-grained variations in the images, exhibiting superior performance compared to other learned growth models.
Deep Probabilistic Modeling of Glioma Growth
Jul 09, 2019


Abstract:Existing approaches to modeling the dynamics of brain tumor growth, specifically glioma, employ biologically inspired models of cell diffusion, using image data to estimate the associated parameters. In this work, we propose an alternative approach based on recent advances in probabilistic segmentation and representation learning that implicitly learns growth dynamics directly from data without an underlying explicit model. We present evidence that our approach is able to learn a distribution of plausible future tumor appearances conditioned on past observations of the same tumor.
Automated brain extraction of multi-sequence MRI using artificial neural networks
Jan 31, 2019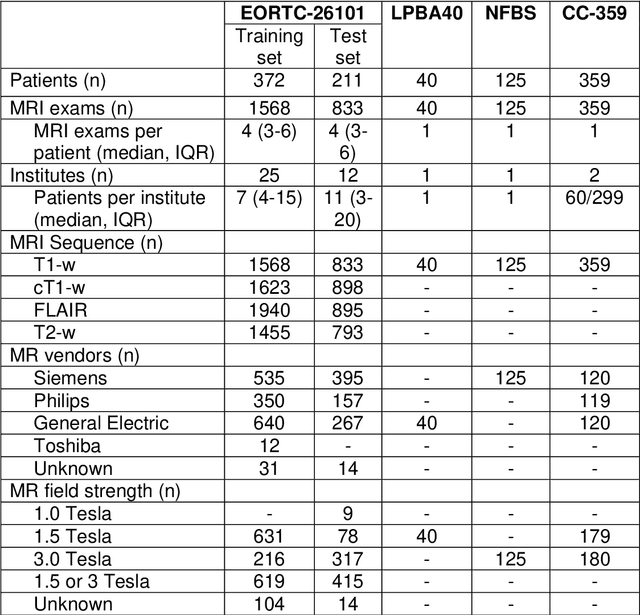
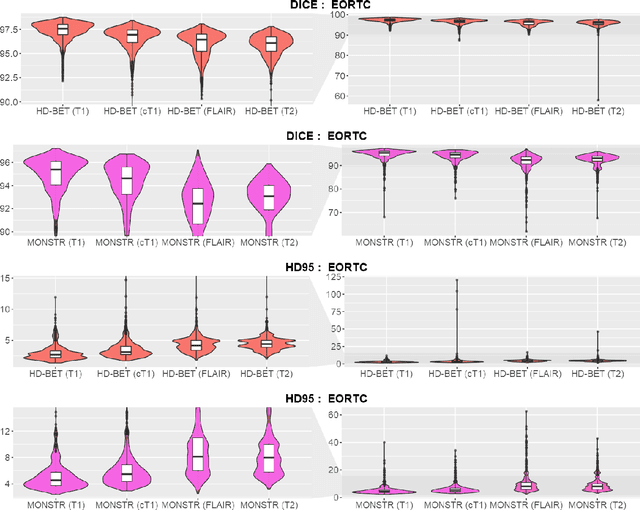
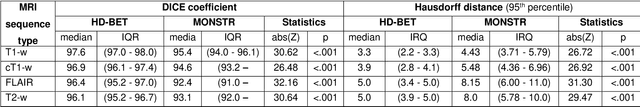
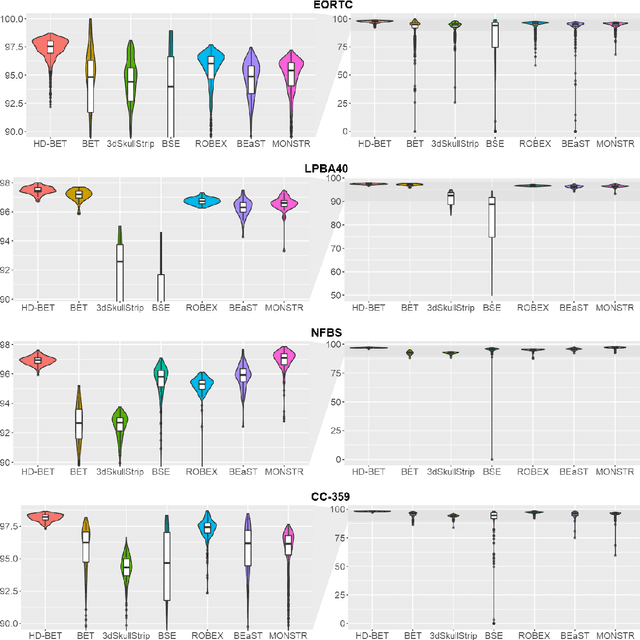
Abstract:Brain extraction is a critical preprocessing step in the analysis of MRI neuroimaging studies and influences the accuracy of downstream analyses. State-of-the-art brain extraction algorithms are, however, optimized for processing healthy brains and thus frequently fail in the presence of pathologically altered brain or when applied to heterogeneous MRI datasets. Here we introduce a new, rigorously validated algorithm (termed HD-BET) relying on artificial neural networks that aims to overcome these limitations. We demonstrate that HD-BET outperforms five publicly available state-of-the-art brain extraction algorithms in several large-scale neuroimaging datasets, including one from a prospective multicentric trial in neuro-oncology, yielding median improvements of +1.33 to +2.63 points for the DICE coefficient and -0.80 to -2.75 mm for the Hausdorff distance (Bonferroni-adjusted p<0.001). Importantly, the HD-BET algorithm shows robust performance in the presence of pathology or treatment-induced tissue alterations, is applicable to a broad range of MRI sequence types and is not influenced by variations in MRI hardware and acquisition parameters encountered in both research and clinical practice. For broader accessibility our HD-BET prediction algorithm is made freely available and may become an essential component for robust, automated, high-throughput processing of MRI neuroimaging data.
No New-Net
Sep 27, 2018
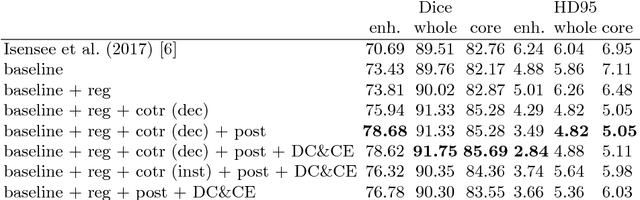
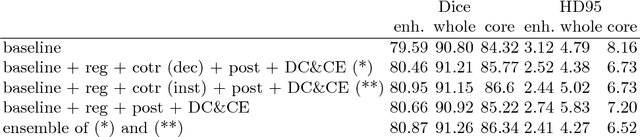

Abstract:In this paper we demonstrate the effectiveness of a well trained U-Net in the context of the BraTS 2018 challenge. This endeavour is particularly interesting given that researchers are currently besting each other with architectural modifications that are intended to improve the segmentation performance. We instead focus on the training process, argue that a well trained U-Net is hard to beat and intend to back up this assumption with a strong participation in this years BraTS challenge. Our baseline U-Net, which has only minor modifications and is trained with a large patch size and a dice loss function already achieves competitive dice scores on the BraTS2018 validation data. By incorporating region based training, additional training data and a simple postprocessing technique, we obtain dice scores of 81.01, 90.83 and 85.44 and Hausdorff Distances (95th percentile) of 2.54, 4.97 and 7.
Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge
Feb 28, 2018
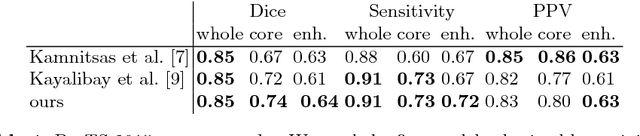

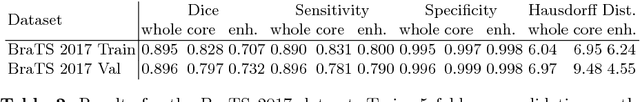
Abstract:Quantitative analysis of brain tumors is critical for clinical decision making. While manual segmentation is tedious, time consuming and subjective, this task is at the same time very challenging to solve for automatic segmentation methods. In this paper we present our most recent effort on developing a robust segmentation algorithm in the form of a convolutional neural network. Our network architecture was inspired by the popular U-Net and has been carefully modified to maximize brain tumor segmentation performance. We use a dice loss function to cope with class imbalances and use extensive data augmentation to successfully prevent overfitting. Our method beats the current state of the art on BraTS 2015, is one of the leading methods on the BraTS 2017 validation set (dice scores of 0.896, 0.797 and 0.732 for whole tumor, tumor core and enhancing tumor, respectively) and achieves very good Dice scores on the test set (0.858 for whole, 0.775 for core and 0.647 for enhancing tumor). We furthermore take part in the survival prediction subchallenge by training an ensemble of a random forest regressor and multilayer perceptrons on shape features describing the tumor subregions. Our approach achieves 52.6% accuracy, a Spearman correlation coefficient of 0.496 and a mean square error of 209607 on the test set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge