Jisoo Lee
Mi:dm 2.0 Korea-centric Bilingual Language Models
Jan 14, 2026Abstract:We introduce Mi:dm 2.0, a bilingual large language model (LLM) specifically engineered to advance Korea-centric AI. This model goes beyond Korean text processing by integrating the values, reasoning patterns, and commonsense knowledge inherent to Korean society, enabling nuanced understanding of cultural contexts, emotional subtleties, and real-world scenarios to generate reliable and culturally appropriate responses. To address limitations of existing LLMs, often caused by insufficient or low-quality Korean data and lack of cultural alignment, Mi:dm 2.0 emphasizes robust data quality through a comprehensive pipeline that includes proprietary data cleansing, high-quality synthetic data generation, strategic data mixing with curriculum learning, and a custom Korean-optimized tokenizer to improve efficiency and coverage. To realize this vision, we offer two complementary configurations: Mi:dm 2.0 Base (11.5B parameters), built with a depth-up scaling strategy for general-purpose use, and Mi:dm 2.0 Mini (2.3B parameters), optimized for resource-constrained environments and specialized tasks. Mi:dm 2.0 achieves state-of-the-art performance on Korean-specific benchmarks, with top-tier zero-shot results on KMMLU and strong internal evaluation results across language, humanities, and social science tasks. The Mi:dm 2.0 lineup is released under the MIT license to support extensive research and commercial use. By offering accessible and high-performance Korea-centric LLMs, KT aims to accelerate AI adoption across Korean industries, public services, and education, strengthen the Korean AI developer community, and lay the groundwork for the broader vision of K-intelligence. Our models are available at https://huggingface.co/K-intelligence. For technical inquiries, please contact midm-llm@kt.com.
Reliable Grid Forecasting: State Space Models for Safety-Critical Energy Systems
Jan 08, 2026Abstract:Accurate grid load forecasting is safety-critical: under-predictions risk supply shortfalls, while symmetric error metrics mask this operational asymmetry. We introduce a grid-specific evaluation framework (Asymmetric MAPE, Under-Prediction Rate, and Reserve Margin) that directly measures operational risk rather than statistical accuracy alone. Using this framework, we conduct a systematic evaluation of Mamba-based State Space Models for California grid forecasting on a weather-aligned CA ISO-TAC dataset spanning Nov 2023 to Nov 2025 (84,498 hourly records across 5 transmission areas). Our analysis reveals that standard accuracy metrics are poor proxies for operational safety: models with identical MAPE can require vastly different reserve margins. We demonstrate that forecast errors are weakly but statistically significantly associated with temperature (r = 0.16), motivating weather-aware modeling rather than loss function modification alone. The S-Mamba model achieves the lowest 99.5th-percentile reserve margin (14.12 percent) compared to 16.66 percent for iTransformer, demonstrating superior forecast reliability under a 99.5th-percentile tail-risk reserve proxy.
Transplant-Ready? Evaluating AI Lung Segmentation Models in Candidates with Severe Lung Disease
Sep 18, 2025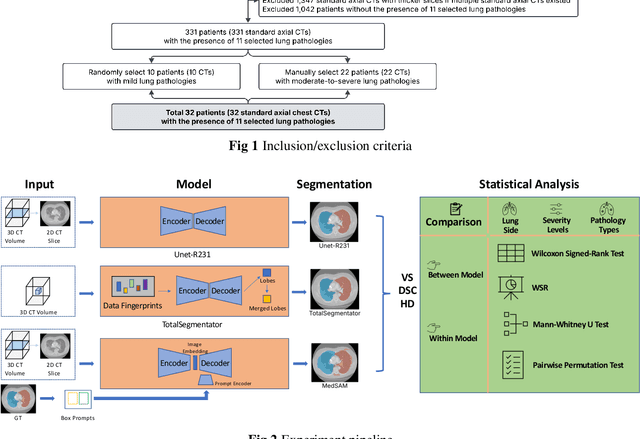
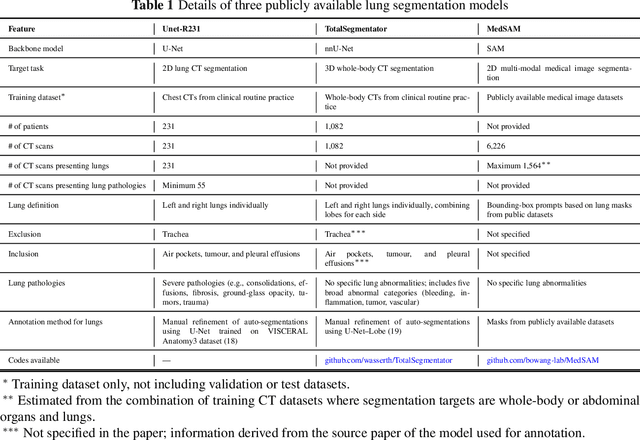
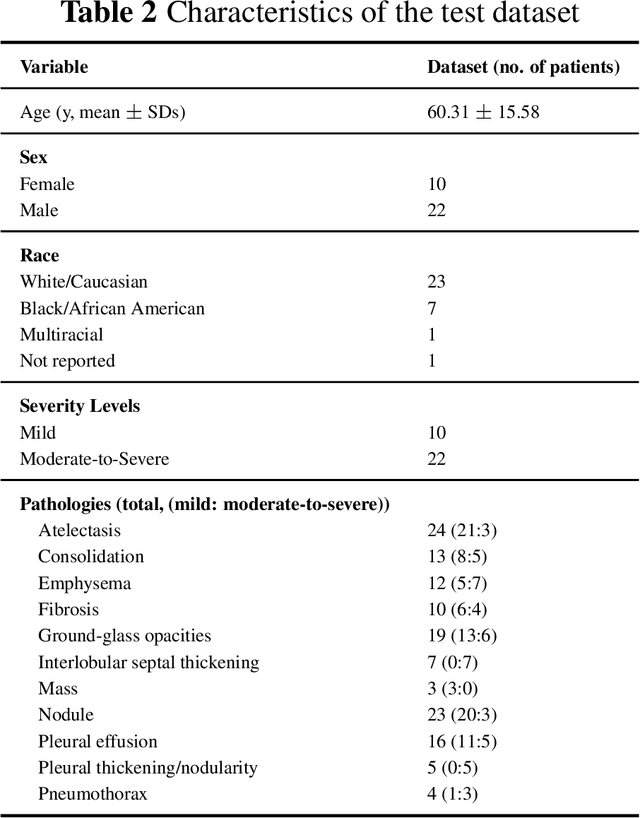
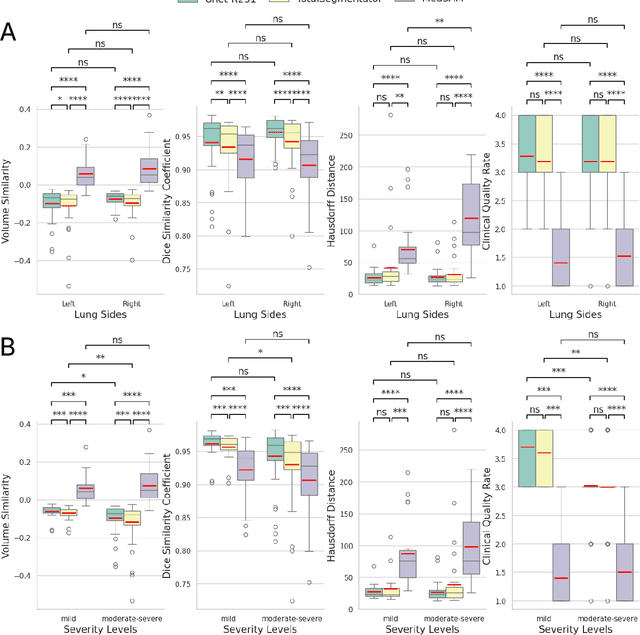
Abstract:This study evaluates publicly available deep-learning based lung segmentation models in transplant-eligible patients to determine their performance across disease severity levels, pathology categories, and lung sides, and to identify limitations impacting their use in preoperative planning in lung transplantation. This retrospective study included 32 patients who underwent chest CT scans at Duke University Health System between 2017 and 2019 (total of 3,645 2D axial slices). Patients with standard axial CT scans were selected based on the presence of two or more lung pathologies of varying severity. Lung segmentation was performed using three previously developed deep learning models: Unet-R231, TotalSegmentator, MedSAM. Performance was assessed using quantitative metrics (volumetric similarity, Dice similarity coefficient, Hausdorff distance) and a qualitative measure (four-point clinical acceptability scale). Unet-R231 consistently outperformed TotalSegmentator and MedSAM in general, for different severity levels, and pathology categories (p<0.05). All models showed significant performance declines from mild to moderate-to-severe cases, particularly in volumetric similarity (p<0.05), without significant differences among lung sides or pathology types. Unet-R231 provided the most accurate automated lung segmentation among evaluated models with TotalSegmentator being a close second, though their performance declined significantly in moderate-to-severe cases, emphasizing the need for specialized model fine-tuning in severe pathology contexts.
MORE-CLEAR: Multimodal Offline Reinforcement learning for Clinical notes Leveraged Enhanced State Representation
Aug 11, 2025



Abstract:Sepsis, a life-threatening inflammatory response to infection, causes organ dysfunction, making early detection and optimal management critical. Previous reinforcement learning (RL) approaches to sepsis management rely primarily on structured data, such as lab results or vital signs, and on a dearth of a comprehensive understanding of the patient's condition. In this work, we propose a Multimodal Offline REinforcement learning for Clinical notes Leveraged Enhanced stAte Representation (MORE-CLEAR) framework for sepsis control in intensive care units. MORE-CLEAR employs pre-trained large-scale language models (LLMs) to facilitate the extraction of rich semantic representations from clinical notes, preserving clinical context and improving patient state representation. Gated fusion and cross-modal attention allow dynamic weight adjustment in the context of time and the effective integration of multimodal data. Extensive cross-validation using two public (MIMIC-III and MIMIC-IV) and one private dataset demonstrates that MORE-CLEAR significantly improves estimated survival rate and policy performance compared to single-modal RL approaches. To our knowledge, this is the first to leverage LLM capabilities within a multimodal offline RL for better state representation in medical applications. This approach can potentially expedite the treatment and management of sepsis by enabling reinforcement learning models to propose enhanced actions based on a more comprehensive understanding of patient conditions.
GEMMAS: Graph-based Evaluation Metrics for Multi Agent Systems
Jul 17, 2025Abstract:Multi-agent systems built on language models have shown strong performance on collaborative reasoning tasks. However, existing evaluations focus only on the correctness of the final output, overlooking how inefficient communication and poor coordination contribute to redundant reasoning and higher computational costs. We introduce GEMMAS, a graph-based evaluation framework that analyzes the internal collaboration process by modeling agent interactions as a directed acyclic graph. To capture collaboration quality, we propose two process-level metrics: Information Diversity Score (IDS) to measure semantic variation in inter-agent messages, and Unnecessary Path Ratio (UPR) to quantify redundant reasoning paths. We evaluate GEMMAS across five benchmarks and highlight results on GSM8K, where systems with only a 2.1% difference in accuracy differ by 12.8% in IDS and 80% in UPR, revealing substantial variation in internal collaboration. These findings demonstrate that outcome-only metrics are insufficient for evaluating multi-agent performance and highlight the importance of process-level diagnostics in designing more interpretable and resource-efficient collaborative AI systems.
Improving Surgical Risk Prediction Through Integrating Automated Body Composition Analysis: a Retrospective Trial on Colectomy Surgery
Jun 16, 2025Abstract:Objective: To evaluate whether preoperative body composition metrics automatically extracted from CT scans can predict postoperative outcomes after colectomy, either alone or combined with clinical variables or existing risk predictors. Main outcomes and measures: The primary outcome was the predictive performance for 1-year all-cause mortality following colectomy. A Cox proportional hazards model with 1-year follow-up was used, and performance was evaluated using the concordance index (C-index) and Integrated Brier Score (IBS). Secondary outcomes included postoperative complications, unplanned readmission, blood transfusion, and severe infection, assessed using AUC and Brier Score from logistic regression. Odds ratios (OR) described associations between individual CT-derived body composition metrics and outcomes. Over 300 features were extracted from preoperative CTs across multiple vertebral levels, including skeletal muscle area, density, fat areas, and inter-tissue metrics. NSQIP scores were available for all surgeries after 2012.
Ground Reaction Force Estimation via Time-aware Knowledge Distillation
Jun 12, 2025Abstract:Human gait analysis with wearable sensors has been widely used in various applications, such as daily life healthcare, rehabilitation, physical therapy, and clinical diagnostics and monitoring. In particular, ground reaction force (GRF) provides critical information about how the body interacts with the ground during locomotion. Although instrumented treadmills have been widely used as the gold standard for measuring GRF during walking, their lack of portability and high cost make them impractical for many applications. As an alternative, low-cost, portable, wearable insole sensors have been utilized to measure GRF; however, these sensors are susceptible to noise and disturbance and are less accurate than treadmill measurements. To address these challenges, we propose a Time-aware Knowledge Distillation framework for GRF estimation from insole sensor data. This framework leverages similarity and temporal features within a mini-batch during the knowledge distillation process, effectively capturing the complementary relationships between features and the sequential properties of the target and input data. The performance of the lightweight models distilled through this framework was evaluated by comparing GRF estimations from insole sensor data against measurements from an instrumented treadmill. Empirical results demonstrated that Time-aware Knowledge Distillation outperforms current baselines in GRF estimation from wearable sensor data.
Automatic Temporal Segmentation for Post-Stroke Rehabilitation: A Keypoint Detection and Temporal Segmentation Approach for Small Datasets
Feb 27, 2025Abstract:Rehabilitation is essential and critical for post-stroke patients, addressing both physical and cognitive aspects. Stroke predominantly affects older adults, with 75% of cases occurring in individuals aged 65 and older, underscoring the urgent need for tailored rehabilitation strategies in aging populations. Despite the critical role therapists play in evaluating rehabilitation progress and ensuring the effectiveness of treatment, current assessment methods can often be subjective, inconsistent, and time-consuming, leading to delays in adjusting therapy protocols. This study aims to address these challenges by providing a solution for consistent and timely analysis. Specifically, we perform temporal segmentation of video recordings to capture detailed activities during stroke patients' rehabilitation. The main application scenario motivating this study is the clinical assessment of daily tabletop object interactions, which are crucial for post-stroke physical rehabilitation. To achieve this, we present a framework that leverages the biomechanics of movement during therapy sessions. Our solution divides the process into two main tasks: 2D keypoint detection to track patients' physical movements, and 1D time-series temporal segmentation to analyze these movements over time. This dual approach enables automated labeling with only a limited set of real-world data, addressing the challenges of variability in patient movements and limited dataset availability. By tackling these issues, our method shows strong potential for practical deployment in physical therapy settings, enhancing the speed and accuracy of rehabilitation assessments.
HyperCLOVA X Technical Report
Apr 13, 2024Abstract:We introduce HyperCLOVA X, a family of large language models (LLMs) tailored to the Korean language and culture, along with competitive capabilities in English, math, and coding. HyperCLOVA X was trained on a balanced mix of Korean, English, and code data, followed by instruction-tuning with high-quality human-annotated datasets while abiding by strict safety guidelines reflecting our commitment to responsible AI. The model is evaluated across various benchmarks, including comprehensive reasoning, knowledge, commonsense, factuality, coding, math, chatting, instruction-following, and harmlessness, in both Korean and English. HyperCLOVA X exhibits strong reasoning capabilities in Korean backed by a deep understanding of the language and cultural nuances. Further analysis of the inherent bilingual nature and its extension to multilingualism highlights the model's cross-lingual proficiency and strong generalization ability to untargeted languages, including machine translation between several language pairs and cross-lingual inference tasks. We believe that HyperCLOVA X can provide helpful guidance for regions or countries in developing their sovereign LLMs.
ContourDiff: Unpaired Image Translation with Contour-Guided Diffusion Models
Mar 16, 2024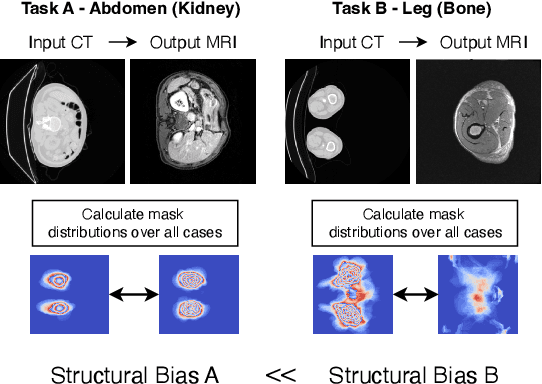
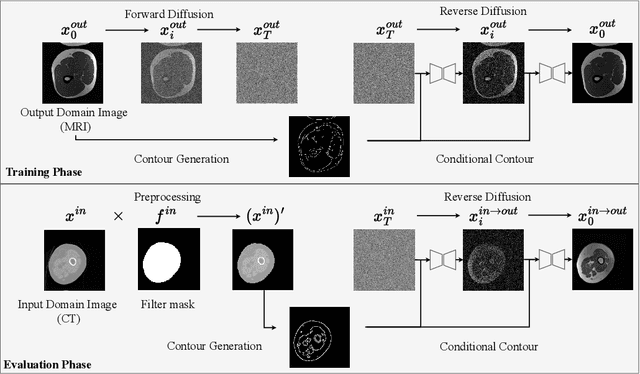
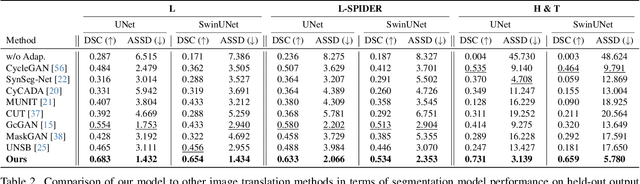
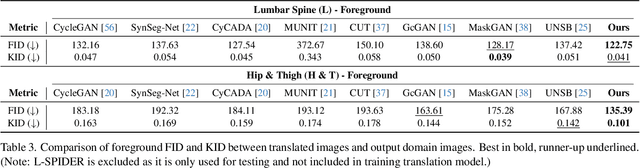
Abstract:Accurately translating medical images across different modalities (e.g., CT to MRI) has numerous downstream clinical and machine learning applications. While several methods have been proposed to achieve this, they often prioritize perceptual quality with respect to output domain features over preserving anatomical fidelity. However, maintaining anatomy during translation is essential for many tasks, e.g., when leveraging masks from the input domain to develop a segmentation model with images translated to the output domain. To address these challenges, we propose ContourDiff, a novel framework that leverages domain-invariant anatomical contour representations of images. These representations are simple to extract from images, yet form precise spatial constraints on their anatomical content. We introduce a diffusion model that converts contour representations of images from arbitrary input domains into images in the output domain of interest. By applying the contour as a constraint at every diffusion sampling step, we ensure the preservation of anatomical content. We evaluate our method by training a segmentation model on images translated from CT to MRI with their original CT masks and testing its performance on real MRIs. Our method outperforms other unpaired image translation methods by a significant margin, furthermore without the need to access any input domain information during training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge