Kyung-Su Kim
Comparative Validation of AI and non-AI Methods in MRI Volumetry to Diagnose Parkinsonian Syndromes
Jul 23, 2022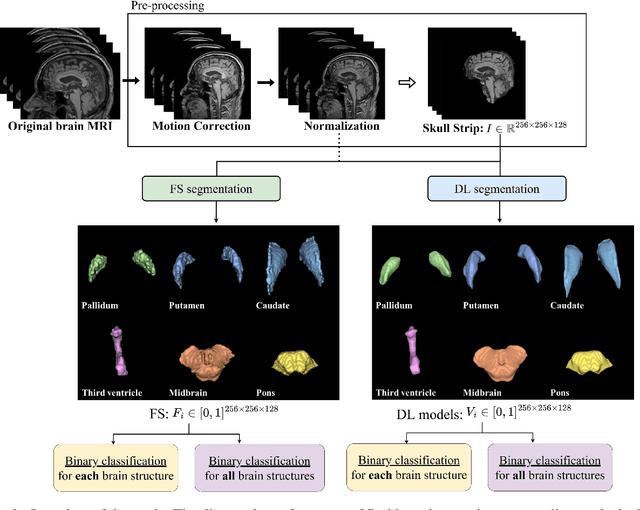

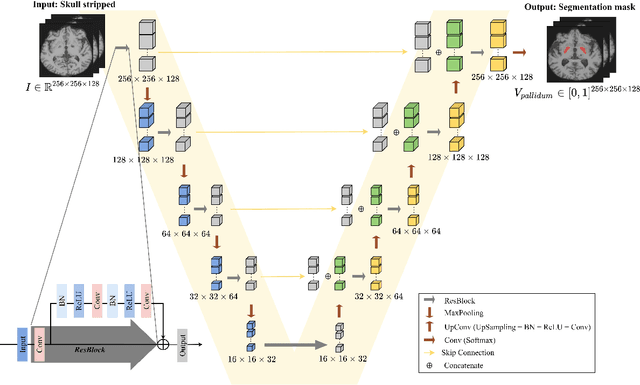
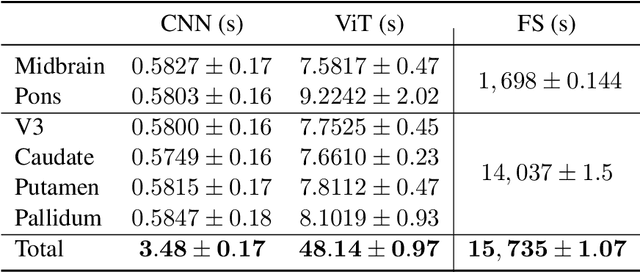
Abstract:Automated segmentation and volumetry of brain magnetic resonance imaging (MRI) scans are essential for the diagnosis of Parkinson's disease (PD) and Parkinson's plus syndromes (P-plus). To enhance the diagnostic performance, we adopt deep learning (DL) models in brain segmentation and compared their performance with the gold-standard non-DL method. We collected brain MRI scans of healthy controls (n=105) and patients with PD (n=105), multiple systemic atrophy (n=132), and progressive supranuclear palsy (n=69) at Samsung Medical Center from January 2017 to December 2020. Using the gold-standard non-DL model, FreeSurfer (FS), we segmented six brain structures: midbrain, pons, caudate, putamen, pallidum, and third ventricle, and considered them as annotating data for DL models, the representative V-Net and UNETR. The Dice scores and area under the curve (AUC) for differentiating normal, PD, and P-plus cases were calculated. The segmentation times of V-Net and UNETR for the six brain structures per patient were 3.48 +- 0.17 and 48.14 +- 0.97 s, respectively, being at least 300 times faster than FS (15,735 +- 1.07 s). Dice scores of both DL models were sufficiently high (>0.85), and their AUCs for disease classification were superior to that of FS. For classification of normal vs. P-plus and PD vs. multiple systemic atrophy (cerebellar type), the DL models and FS showed AUCs above 0.8. DL significantly reduces the analysis time without compromising the performance of brain segmentation and differential diagnosis. Our findings may contribute to the adoption of DL brain MRI segmentation in clinical settings and advance brain research.
Improved Generative Model for Weakly Supervised Chest Anomaly Localization via Pseudo-paired Registration with Bilaterally Symmetrical Data Augmentation
Jul 21, 2022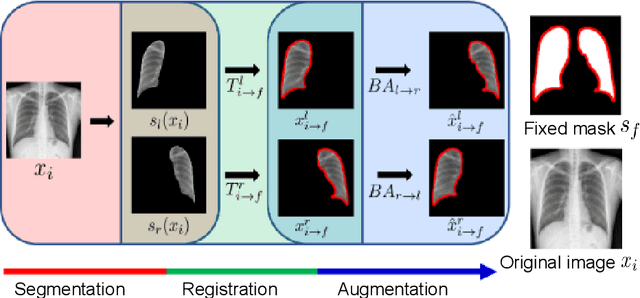
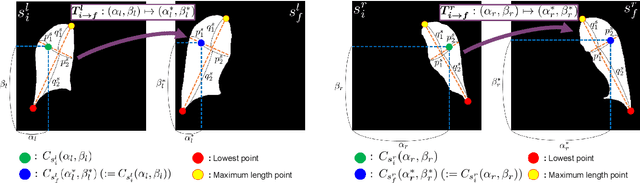
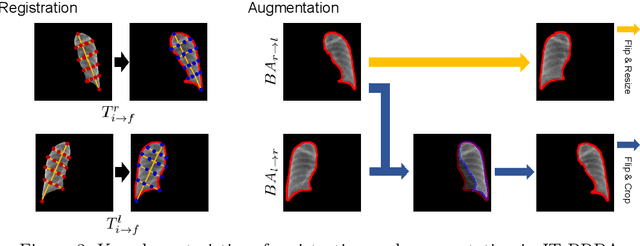
Abstract:Image translation based on a generative adversarial network (GAN-IT) is a promising method for precise localization of abnormal regions in chest X-ray images (AL-CXR). However, heterogeneous unpaired datasets undermine existing methods to extract key features and distinguish normal from abnormal cases, resulting in inaccurate and unstable AL-CXR. To address this problem, we propose an improved two-stage GAN-IT involving registration and data augmentation. For the first stage, we introduce an invertible deep-learning-based registration technique that virtually and reasonably converts unpaired data into paired data for learning registration maps. This novel approach achieves high registration performance. For the second stage, we apply data augmentation to diversify anomaly locations by swapping the left and right lung regions on the uniform registered frames, further improving the performance by alleviating imbalance in data distribution showing left and right lung lesions. Our method is intended for application to existing GAN-IT models, allowing existing architecture to benefit from key features for translation. By showing that the AL-CXR performance is uniformly improved when applying the proposed method, we believe that GAN-IT for AL-CXR can be deployed in clinical environments, even if learning data are scarce.
AI-based computer-aided diagnostic system of chest digital tomography synthesis: Demonstrating comparative advantage with X-ray-based AI systems
Jun 18, 2022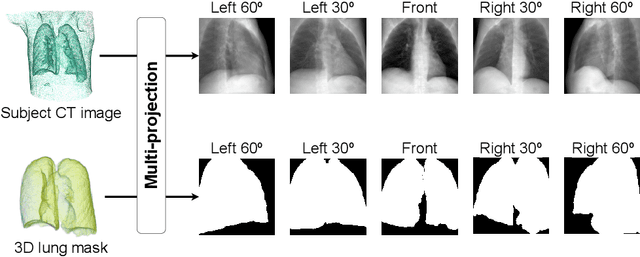
Abstract:Compared with chest X-ray (CXR) imaging, which is a single image projected from the front of the patient, chest digital tomosynthesis (CDTS) imaging can be more advantageous for lung lesion detection because it acquires multiple images projected from multiple angles of the patient. Various clinical comparative analysis and verification studies have been reported to demonstrate this, but there were no artificial intelligence (AI)-based comparative analysis studies. Existing AI-based computer-aided detection (CAD) systems for lung lesion diagnosis have been developed mainly based on CXR images; however, CAD-based on CDTS, which uses multi-angle images of patients in various directions, has not been proposed and verified for its usefulness compared to CXR-based counterparts. This study develops/tests a CDTS-based AI CAD system to detect lung lesions to demonstrate performance improvements compared to CXR-based AI CAD. We used multiple projection images as input for the CDTS-based AI model and a single-projection image as input for the CXR-based AI model to fairly compare and evaluate the performance between models. The proposed CDTS-based AI CAD system yielded sensitivities of 0.782 and 0.785 and accuracies of 0.895 and 0.837 for the performance of detecting tuberculosis and pneumonia, respectively, against normal subjects. These results show higher performance than sensitivities of 0.728 and 0.698 and accuracies of 0.874 and 0.826 for detecting tuberculosis and pneumonia through the CXR-based AI CAD, which only uses a single projection image in the frontal direction. We found that CDTS-based AI CAD improved the sensitivity of tuberculosis and pneumonia by 5.4% and 8.7% respectively, compared to CXR-based AI CAD without loss of accuracy. Therefore, we comparatively prove that CDTS-based AI CAD technology can improve performance more than CXR, enhancing the clinical applicability of CDTS.
3D unsupervised anomaly detection and localization through virtual multi-view projection and reconstruction: Clinical validation on low-dose chest computed tomography
Jun 18, 2022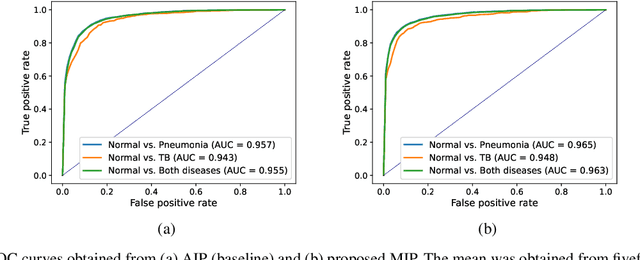
Abstract:Computer-aided diagnosis for low-dose computed tomography (CT) based on deep learning has recently attracted attention as a first-line automatic testing tool because of its high accuracy and low radiation exposure. However, existing methods rely on supervised learning, imposing an additional burden to doctors for collecting disease data or annotating spatial labels for network training, consequently hindering their implementation. We propose a method based on a deep neural network for computer-aided diagnosis called virtual multi-view projection and reconstruction for unsupervised anomaly detection. Presumably, this is the first method that only requires data from healthy patients for training to identify three-dimensional (3D) regions containing any anomalies. The method has three key components. Unlike existing computer-aided diagnosis tools that use conventional CT slices as the network input, our method 1) improves the recognition of 3D lung structures by virtually projecting an extracted 3D lung region to obtain two-dimensional (2D) images from diverse views to serve as network inputs, 2) accommodates the input diversity gain for accurate anomaly detection, and 3) achieves 3D anomaly/disease localization through a novel 3D map restoration method using multiple 2D anomaly maps. The proposed method based on unsupervised learning improves the patient-level anomaly detection by 10% (area under the curve, 0.959) compared with a gold standard based on supervised learning (area under the curve, 0.848), and it localizes the anomaly region with 93% accuracy, demonstrating its high performance.
Automated Precision Localization of Peripherally Inserted Central Catheter Tip through Model-Agnostic Multi-Stage Networks
Jun 14, 2022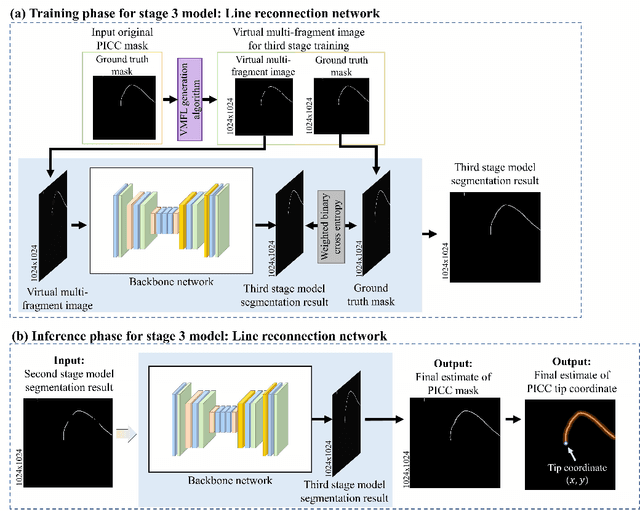

Abstract:Peripherally inserted central catheters (PICCs) have been widely used as one of the representative central venous lines (CVCs) due to their long-term intravascular access with low infectivity. However, PICCs have a fatal drawback of a high frequency of tip mispositions, increasing the risk of puncture, embolism, and complications such as cardiac arrhythmias. To automatically and precisely detect it, various attempts have been made by using the latest deep learning (DL) technologies. However, even with these approaches, it is still practically difficult to determine the tip location because the multiple fragments phenomenon (MFP) occurs in the process of predicting and extracting the PICC line required before predicting the tip. This study aimed to develop a system generally applied to existing models and to restore the PICC line more exactly by removing the MFs of the model output, thereby precisely localizing the actual tip position for detecting its disposition. To achieve this, we proposed a multi-stage DL-based framework post-processing the PICC line extraction result of the existing technology. The performance was compared by each root mean squared error (RMSE) and MFP incidence rate according to whether or not MFCN is applied to five conventional models. In internal validation, when MFCN was applied to the existing single model, MFP was improved by an average of 45%. The RMSE was improved by over 63% from an average of 26.85mm (17.16 to 35.80mm) to 9.72mm (9.37 to 10.98mm). In external validation, when MFCN was applied, the MFP incidence rate decreased by an average of 32% and the RMSE decreased by an average of 65\%. Therefore, by applying the proposed MFCN, we observed the significant/consistent detection performance improvement of PICC tip location compared to the existing model.
RecurSeed and CertainMix for Weakly Supervised Semantic Segmentation
Apr 14, 2022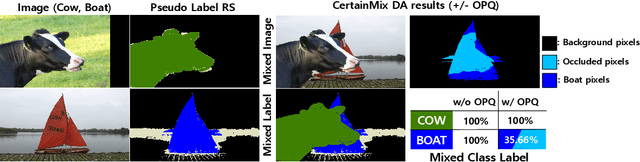
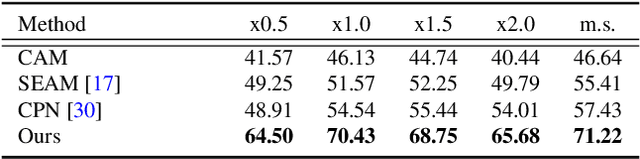
Abstract:Although weakly supervised semantic segmentation using only image-level labels (WSSS-IL) is potentially useful, its low performance and implementation complexity still limit its application. The main causes are (a) non-detection and (b) false-detection phenomena: (a) The class activation maps refined from existing WSSS-IL methods still only represent partial regions for large-scale objects, and (b) for small-scale objects, over-activation causes them to deviate from the object edges. We propose RecurSeed which alternately reduces non- and false-detections through recursive iterations, thereby implicitly finding an optimal junction that minimizes both errors. To maximize the effectiveness of RecurSeed, we also propose a novel data augmentation (DA) approach called CertainMix, which virtually creates object masks and further expresses their edges in combining the segmentation results, thereby obtaining a new DA method effectively reflecting object existence reliability through the spatial information. We achieved new state-of-the-art performances on both the PASCAL VOC 2012 and MS COCO 2014 benchmarks (VOC val 72.4%, COCO val 45.0%). The code is available at https://github.com/OFRIN/RecurSeed_and_CertainMix.
A Revision of Neural Tangent Kernel-based Approaches for Neural Networks
Jul 02, 2020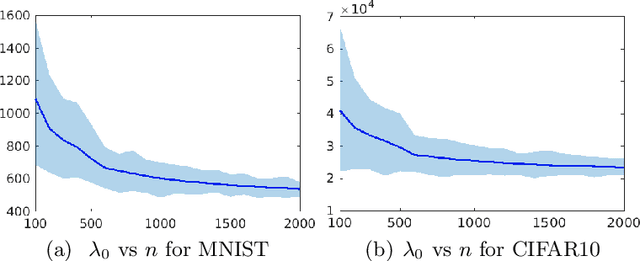
Abstract:Recent theoretical works based on the neural tangent kernel (NTK) have shed light on the optimization and generalization of over-parameterized networks, and partially bridge the gap between their practical success and classical learning theory. Especially, using the NTK-based approach, the following three representative results were obtained: (1) A training error bound was derived to show that networks can fit any finite training sample perfectly by reflecting a tighter characterization of training speed depending on the data complexity. (2) A generalization error bound invariant of network size was derived by using a data-dependent complexity measure (CMD). It follows from this CMD bound that networks can generalize arbitrary smooth functions. (3) A simple and analytic kernel function was derived as indeed equivalent to a fully-trained network. This kernel outperforms its corresponding network and the existing gold standard, Random Forests, in few shot learning. For all of these results to hold, the network scaling factor $\kappa$ should decrease w.r.t. sample size n. In this case of decreasing $\kappa$, however, we prove that the aforementioned results are surprisingly erroneous. It is because the output value of trained network decreases to zero when $\kappa$ decreases w.r.t. n. To solve this problem, we tighten key bounds by essentially removing $\kappa$-affected values. Our tighter analysis resolves the scaling problem and enables the validation of the original NTK-based results.
Compressed Sensing via Measurement-Conditional Generative Models
Jul 02, 2020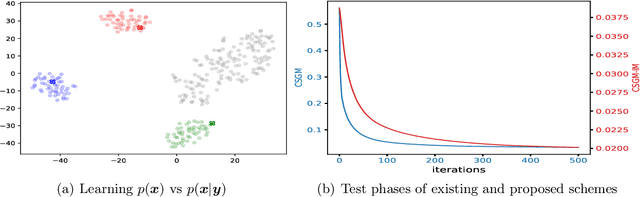
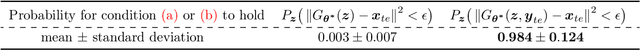
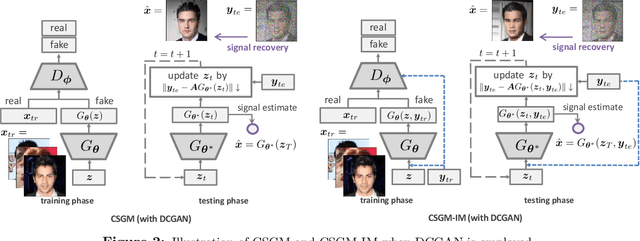
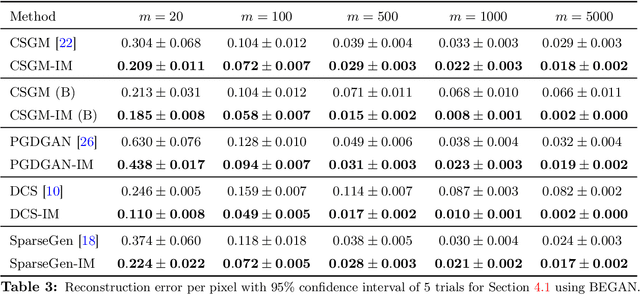
Abstract:A pre-trained generator has been frequently adopted in compressed sensing (CS) due to its ability to effectively estimate signals with the prior of NNs. In order to further refine the NN-based prior, we propose a framework that allows the generator to learn measurement-specific prior distribution, yielding more accurate prediction on a measurement. Our framework has a simple form that only utilizes additional information from a given measurement for prior learning, so it can be easily applied to existing methods. Despite its simplicity, we demonstrate through extensive experiments that our framework exhibits uniformly superior performances by large margin and can reduce the reconstruction error up to an order of magnitude for some applications. We also explain the experimental success in theory by showing that our framework can slightly relax the stringent signal presence condition, which is required to guarantee the success of signal recovery.
Fourier Phase Retrieval with Extended Support Estimation via Deep Neural Network
Apr 03, 2019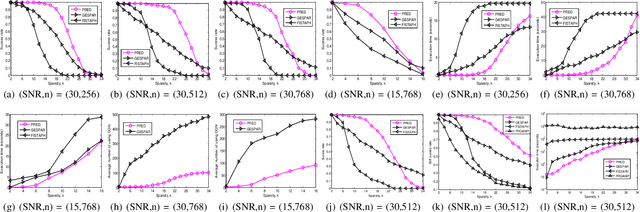
Abstract:We consider the problem of sparse phase retrieval from Fourier transform magnitudes to recover $k$-sparse signal vector $x^{\circ}$ and its support $\mathcal{T}$. To improve the reconstruction performance of $x^{\circ}$, we exploit extended support estimate $\mathcal{E}$ of size larger than $k$ satisfying $\mathcal{E} \supseteq \mathcal{T}$. We propose a learning method for the deep neural network to provide $\mathcal{E}$ as an union of equivalent solutions of $\mathcal{T}$ by utilizing modulo Fourier invariances and suggest a searching technique for $\mathcal{T}$ by iteratively sampling $\mathcal{E}$ from the trained network output and applying the hard thresholding to $\mathcal{E}$. Numerical results show that our proposed scheme has a superior performance with a lower complexity compared to the local search-based greedy sparse phase retrieval method and a state-of-the-art variant of the Fienup method.
Tree Search Network for Sparse Regression
Apr 01, 2019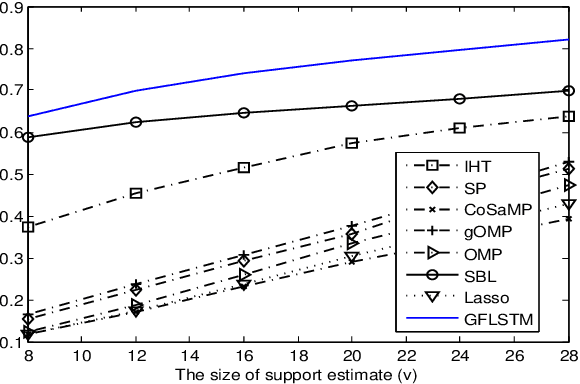
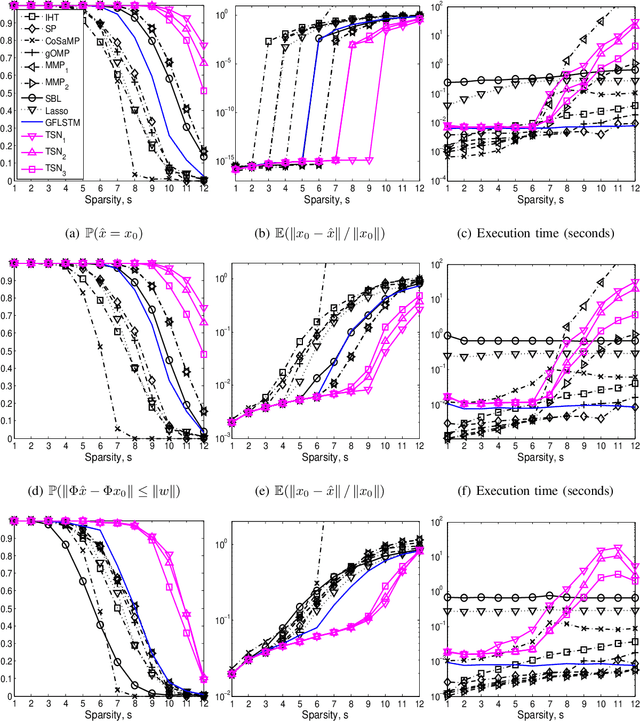
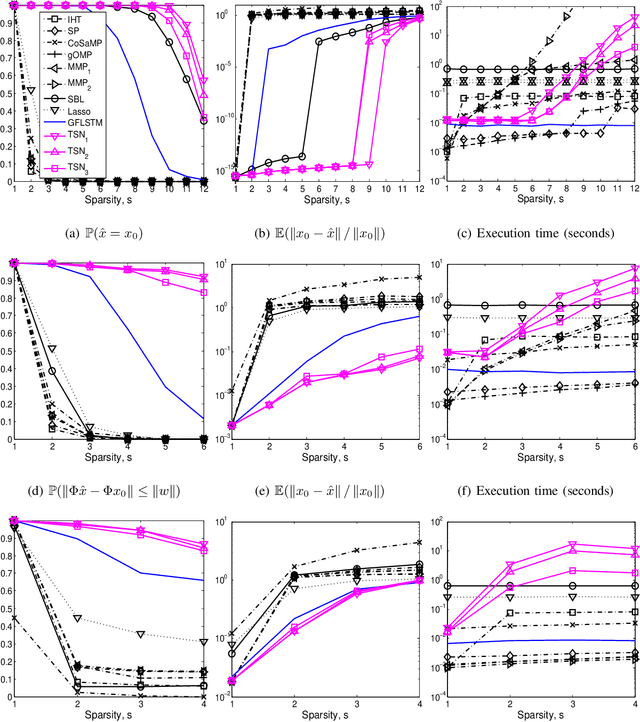
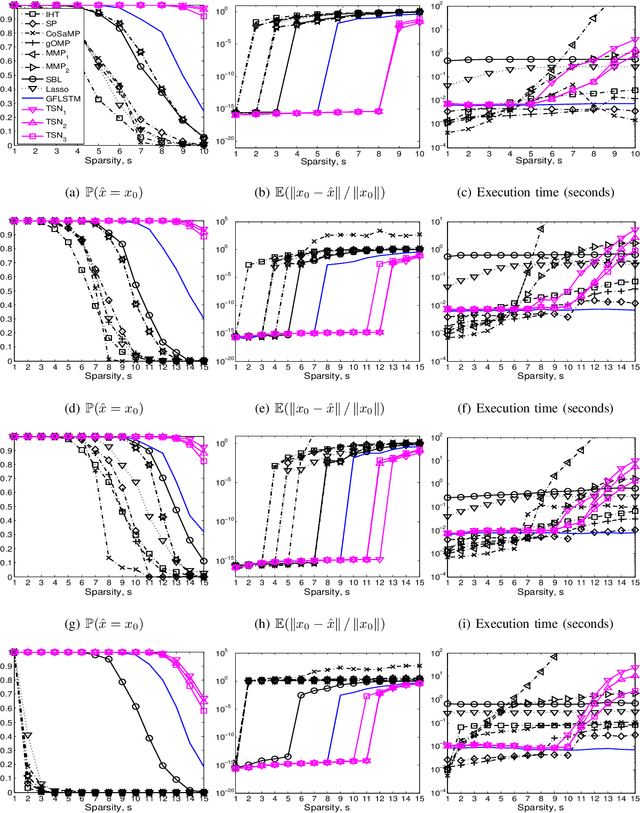
Abstract:We consider the classical sparse regression problem of recovering a sparse signal $x_0$ given a measurement vector $y = \Phi x_0+w$. We propose a tree search algorithm driven by the deep neural network for sparse regression (TSN). TSN improves the signal reconstruction performance of the deep neural network designed for sparse regression by performing a tree search with pruning. It is observed in both noiseless and noisy cases, TSN recovers synthetic and real signals with lower complexity than a conventional tree search and is superior to existing algorithms by a large margin for various types of the sensing matrix $\Phi$, widely used in sparse regression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge