Seong Je Oh
EraseLoRA: MLLM-Driven Foreground Exclusion and Background Subtype Aggregation for Dataset-Free Object Removal
Dec 25, 2025



Abstract:Object removal differs from common inpainting, since it must prevent the masked target from reappearing and reconstruct the occluded background with structural and contextual fidelity, rather than merely filling a hole plausibly. Recent dataset-free approaches that redirect self-attention inside the mask fail in two ways: non-target foregrounds are often misinterpreted as background, which regenerates unwanted objects, and direct attention manipulation disrupts fine details and hinders coherent integration of background cues. We propose EraseLoRA, a novel dataset-free framework that replaces attention surgery with background-aware reasoning and test-time adaptation. First, Background-aware Foreground Exclusion (BFE), uses a multimodal large-language models to separate target foreground, non-target foregrounds, and clean background from a single image-mask pair without paired supervision, producing reliable background cues while excluding distractors. Second, Background-aware Reconstruction with Subtype Aggregation (BRSA), performs test-time optimization that treats inferred background subtypes as complementary pieces and enforces their consistent integration through reconstruction and alignment objectives, preserving local detail and global structure without explicit attention intervention. We validate EraseLoRA as a plug-in to pretrained diffusion models and across benchmarks for object removal, demonstrating consistent improvements over dataset-free baselines and competitive results against dataset-driven methods. The code will be made available upon publication.
Improved Generative Model for Weakly Supervised Chest Anomaly Localization via Pseudo-paired Registration with Bilaterally Symmetrical Data Augmentation
Jul 21, 2022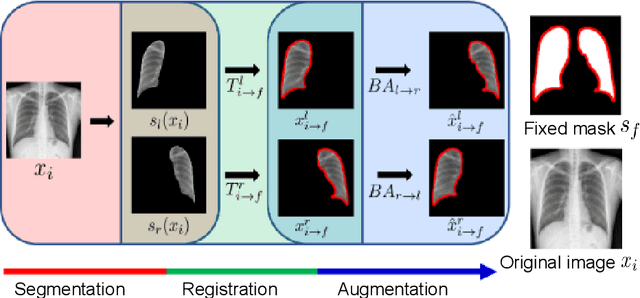
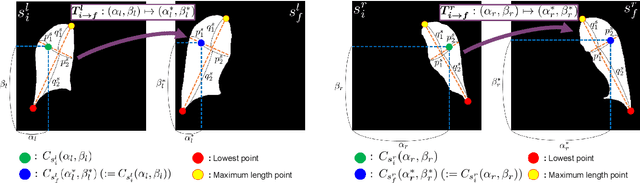
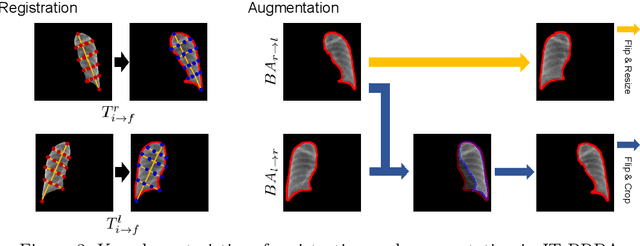
Abstract:Image translation based on a generative adversarial network (GAN-IT) is a promising method for precise localization of abnormal regions in chest X-ray images (AL-CXR). However, heterogeneous unpaired datasets undermine existing methods to extract key features and distinguish normal from abnormal cases, resulting in inaccurate and unstable AL-CXR. To address this problem, we propose an improved two-stage GAN-IT involving registration and data augmentation. For the first stage, we introduce an invertible deep-learning-based registration technique that virtually and reasonably converts unpaired data into paired data for learning registration maps. This novel approach achieves high registration performance. For the second stage, we apply data augmentation to diversify anomaly locations by swapping the left and right lung regions on the uniform registered frames, further improving the performance by alleviating imbalance in data distribution showing left and right lung lesions. Our method is intended for application to existing GAN-IT models, allowing existing architecture to benefit from key features for translation. By showing that the AL-CXR performance is uniformly improved when applying the proposed method, we believe that GAN-IT for AL-CXR can be deployed in clinical environments, even if learning data are scarce.
AI-based computer-aided diagnostic system of chest digital tomography synthesis: Demonstrating comparative advantage with X-ray-based AI systems
Jun 18, 2022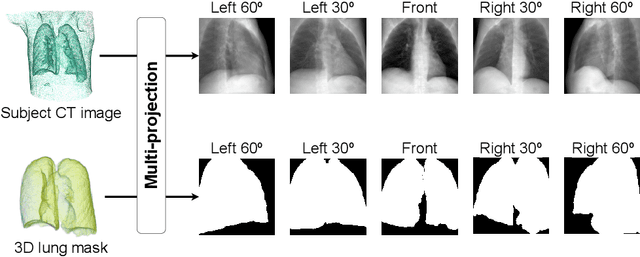
Abstract:Compared with chest X-ray (CXR) imaging, which is a single image projected from the front of the patient, chest digital tomosynthesis (CDTS) imaging can be more advantageous for lung lesion detection because it acquires multiple images projected from multiple angles of the patient. Various clinical comparative analysis and verification studies have been reported to demonstrate this, but there were no artificial intelligence (AI)-based comparative analysis studies. Existing AI-based computer-aided detection (CAD) systems for lung lesion diagnosis have been developed mainly based on CXR images; however, CAD-based on CDTS, which uses multi-angle images of patients in various directions, has not been proposed and verified for its usefulness compared to CXR-based counterparts. This study develops/tests a CDTS-based AI CAD system to detect lung lesions to demonstrate performance improvements compared to CXR-based AI CAD. We used multiple projection images as input for the CDTS-based AI model and a single-projection image as input for the CXR-based AI model to fairly compare and evaluate the performance between models. The proposed CDTS-based AI CAD system yielded sensitivities of 0.782 and 0.785 and accuracies of 0.895 and 0.837 for the performance of detecting tuberculosis and pneumonia, respectively, against normal subjects. These results show higher performance than sensitivities of 0.728 and 0.698 and accuracies of 0.874 and 0.826 for detecting tuberculosis and pneumonia through the CXR-based AI CAD, which only uses a single projection image in the frontal direction. We found that CDTS-based AI CAD improved the sensitivity of tuberculosis and pneumonia by 5.4% and 8.7% respectively, compared to CXR-based AI CAD without loss of accuracy. Therefore, we comparatively prove that CDTS-based AI CAD technology can improve performance more than CXR, enhancing the clinical applicability of CDTS.
3D unsupervised anomaly detection and localization through virtual multi-view projection and reconstruction: Clinical validation on low-dose chest computed tomography
Jun 18, 2022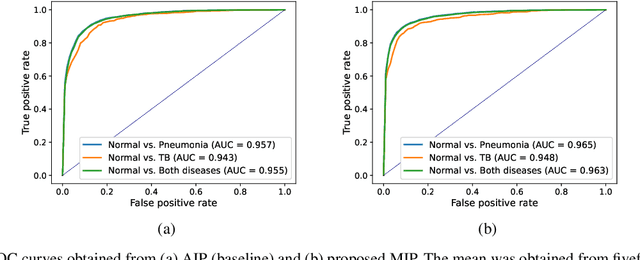
Abstract:Computer-aided diagnosis for low-dose computed tomography (CT) based on deep learning has recently attracted attention as a first-line automatic testing tool because of its high accuracy and low radiation exposure. However, existing methods rely on supervised learning, imposing an additional burden to doctors for collecting disease data or annotating spatial labels for network training, consequently hindering their implementation. We propose a method based on a deep neural network for computer-aided diagnosis called virtual multi-view projection and reconstruction for unsupervised anomaly detection. Presumably, this is the first method that only requires data from healthy patients for training to identify three-dimensional (3D) regions containing any anomalies. The method has three key components. Unlike existing computer-aided diagnosis tools that use conventional CT slices as the network input, our method 1) improves the recognition of 3D lung structures by virtually projecting an extracted 3D lung region to obtain two-dimensional (2D) images from diverse views to serve as network inputs, 2) accommodates the input diversity gain for accurate anomaly detection, and 3) achieves 3D anomaly/disease localization through a novel 3D map restoration method using multiple 2D anomaly maps. The proposed method based on unsupervised learning improves the patient-level anomaly detection by 10% (area under the curve, 0.959) compared with a gold standard based on supervised learning (area under the curve, 0.848), and it localizes the anomaly region with 93% accuracy, demonstrating its high performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge