Yuwen Chen
ETH Zurich
Transplant-Ready? Evaluating AI Lung Segmentation Models in Candidates with Severe Lung Disease
Sep 18, 2025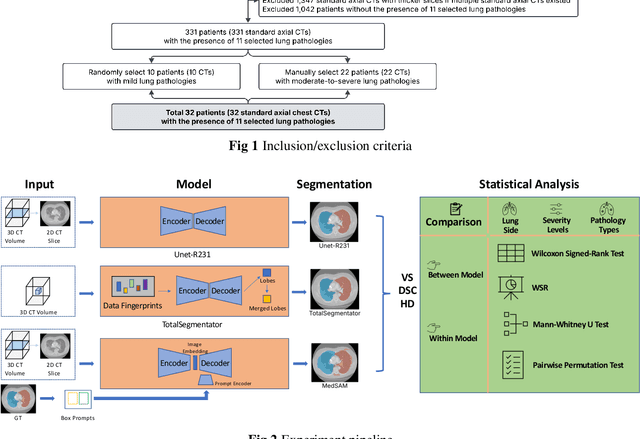
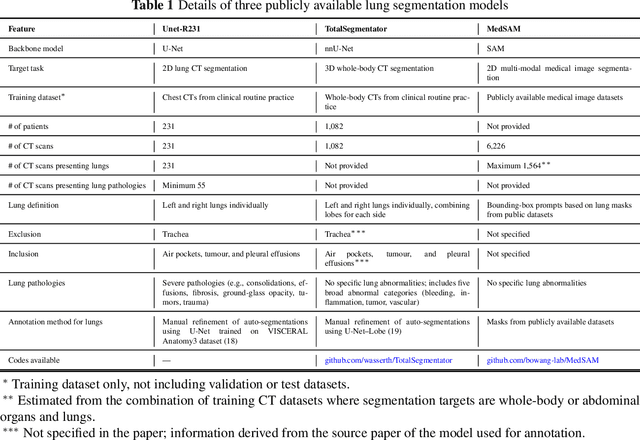
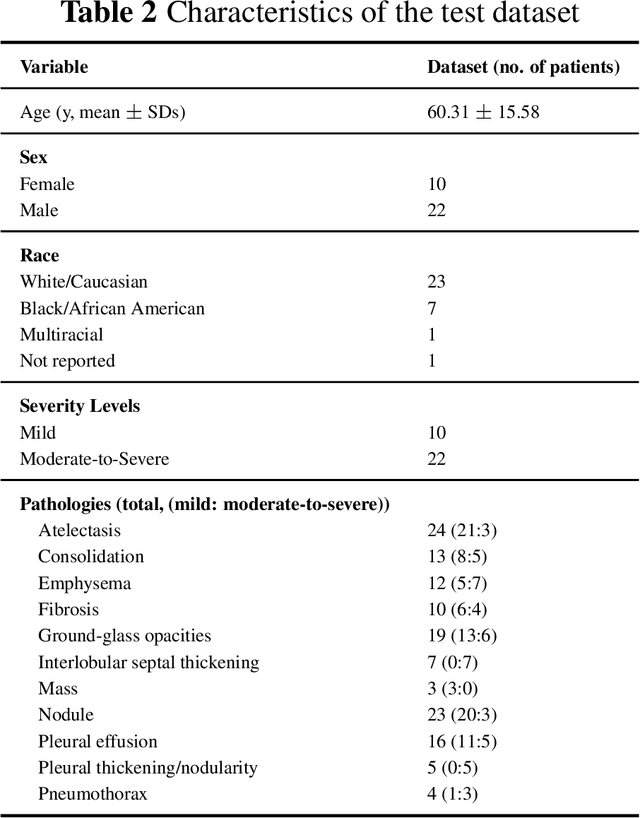
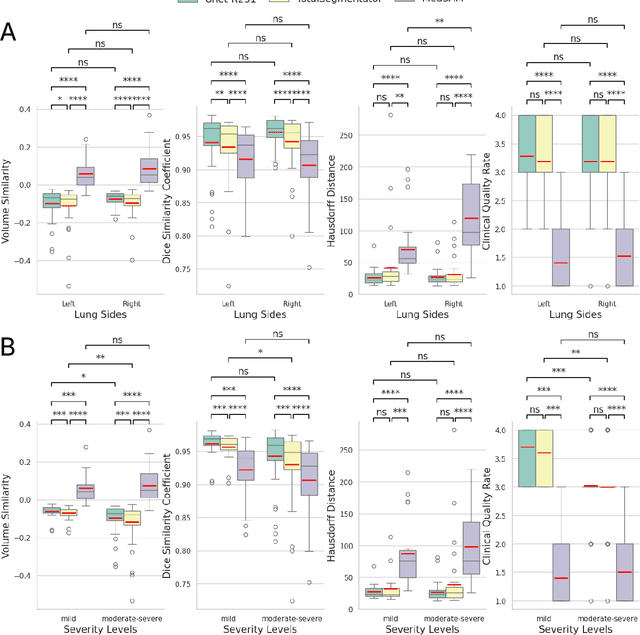
Abstract:This study evaluates publicly available deep-learning based lung segmentation models in transplant-eligible patients to determine their performance across disease severity levels, pathology categories, and lung sides, and to identify limitations impacting their use in preoperative planning in lung transplantation. This retrospective study included 32 patients who underwent chest CT scans at Duke University Health System between 2017 and 2019 (total of 3,645 2D axial slices). Patients with standard axial CT scans were selected based on the presence of two or more lung pathologies of varying severity. Lung segmentation was performed using three previously developed deep learning models: Unet-R231, TotalSegmentator, MedSAM. Performance was assessed using quantitative metrics (volumetric similarity, Dice similarity coefficient, Hausdorff distance) and a qualitative measure (four-point clinical acceptability scale). Unet-R231 consistently outperformed TotalSegmentator and MedSAM in general, for different severity levels, and pathology categories (p<0.05). All models showed significant performance declines from mild to moderate-to-severe cases, particularly in volumetric similarity (p<0.05), without significant differences among lung sides or pathology types. Unet-R231 provided the most accurate automated lung segmentation among evaluated models with TotalSegmentator being a close second, though their performance declined significantly in moderate-to-severe cases, emphasizing the need for specialized model fine-tuning in severe pathology contexts.
BreastSegNet: Multi-label Segmentation of Breast MRI
Jul 18, 2025Abstract:Breast MRI provides high-resolution imaging critical for breast cancer screening and preoperative staging. However, existing segmentation methods for breast MRI remain limited in scope, often focusing on only a few anatomical structures, such as fibroglandular tissue or tumors, and do not cover the full range of tissues seen in scans. This narrows their utility for quantitative analysis. In this study, we present BreastSegNet, a multi-label segmentation algorithm for breast MRI that covers nine anatomical labels: fibroglandular tissue (FGT), vessel, muscle, bone, lesion, lymph node, heart, liver, and implant. We manually annotated a large set of 1123 MRI slices capturing these structures with detailed review and correction from an expert radiologist. Additionally, we benchmark nine segmentation models, including U-Net, SwinUNet, UNet++, SAM, MedSAM, and nnU-Net with multiple ResNet-based encoders. Among them, nnU-Net ResEncM achieves the highest average Dice scores of 0.694 across all labels. It performs especially well on heart, liver, muscle, FGT, and bone, with Dice scores exceeding 0.73, and approaching 0.90 for heart and liver. All model code and weights are publicly available, and we plan to release the data at a later date.
CrowdTrack: A Benchmark for Difficult Multiple Pedestrian Tracking in Real Scenarios
Jul 03, 2025Abstract:Multi-object tracking is a classic field in computer vision. Among them, pedestrian tracking has extremely high application value and has become the most popular research category. Existing methods mainly use motion or appearance information for tracking, which is often difficult in complex scenarios. For the motion information, mutual occlusions between objects often prevent updating of the motion state; for the appearance information, non-robust results are often obtained due to reasons such as only partial visibility of the object or blurred images. Although learning how to perform tracking in these situations from the annotated data is the simplest solution, the existing MOT dataset fails to satisfy this solution. Existing methods mainly have two drawbacks: relatively simple scene composition and non-realistic scenarios. Although some of the video sequences in existing dataset do not have the above-mentioned drawbacks, the number is far from adequate for research purposes. To this end, we propose a difficult large-scale dataset for multi-pedestrian tracking, shot mainly from the first-person view and all from real-life complex scenarios. We name it ``CrowdTrack'' because there are numerous objects in most of the sequences. Our dataset consists of 33 videos, containing a total of 5,185 trajectories. Each object is annotated with a complete bounding box and a unique object ID. The dataset will provide a platform to facilitate the development of algorithms that remain effective in complex situations. We analyzed the dataset comprehensively and tested multiple SOTA models on our dataset. Besides, we analyzed the performance of the foundation models on our dataset. The dataset and project code is released at: https://github.com/loseevaya/CrowdTrack .
MRI-CORE: A Foundation Model for Magnetic Resonance Imaging
Jun 13, 2025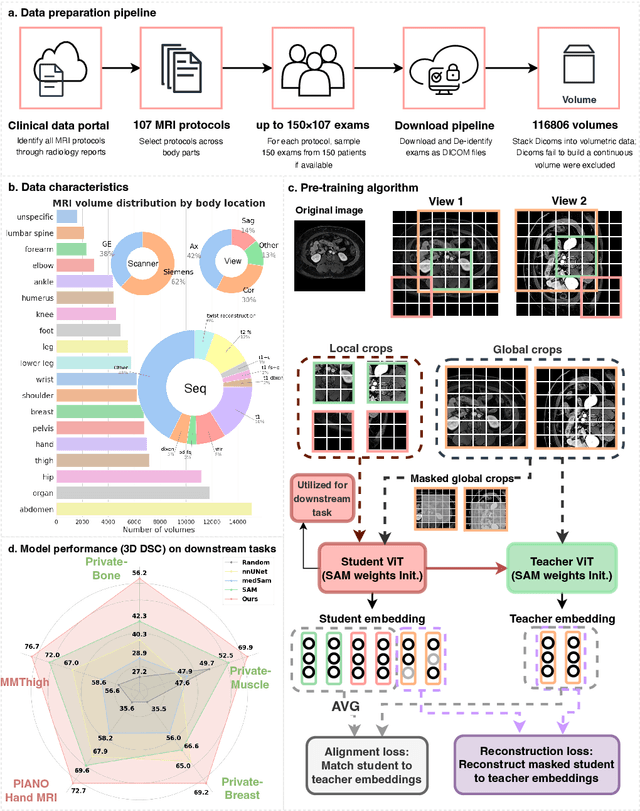
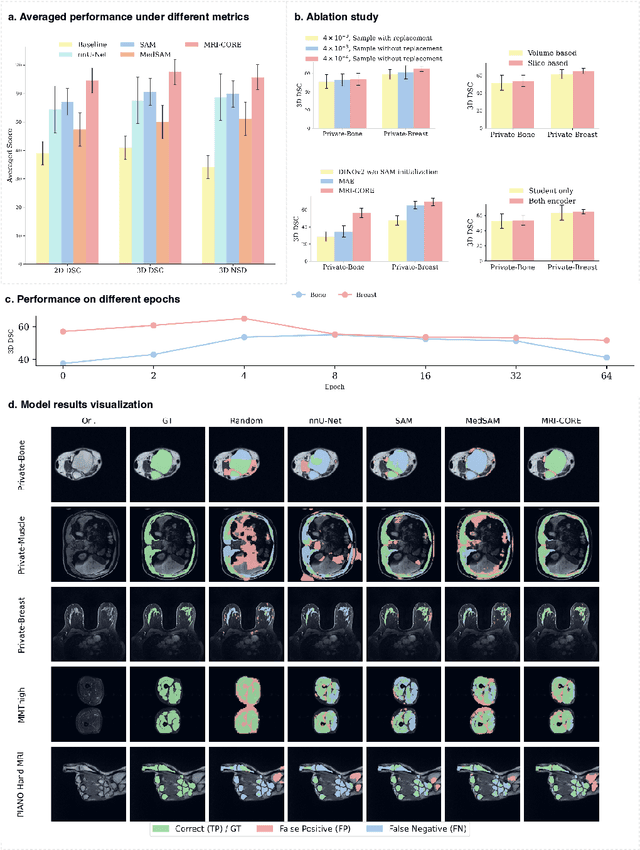
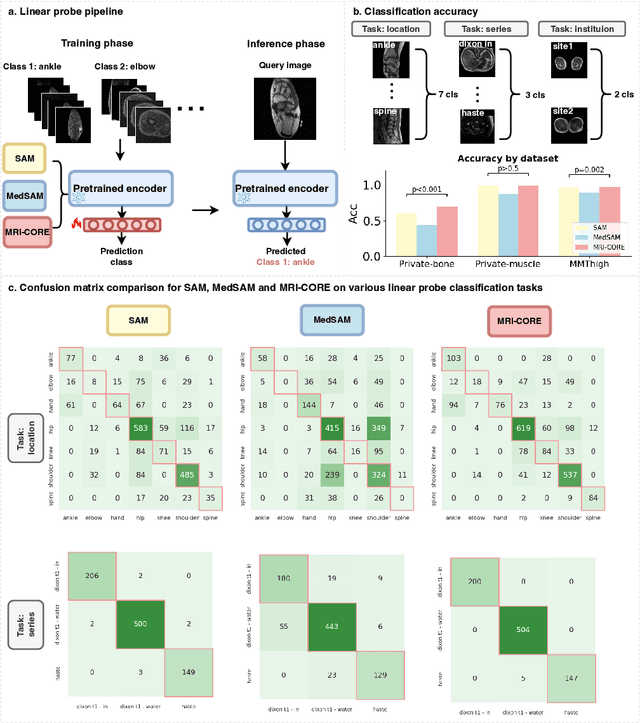
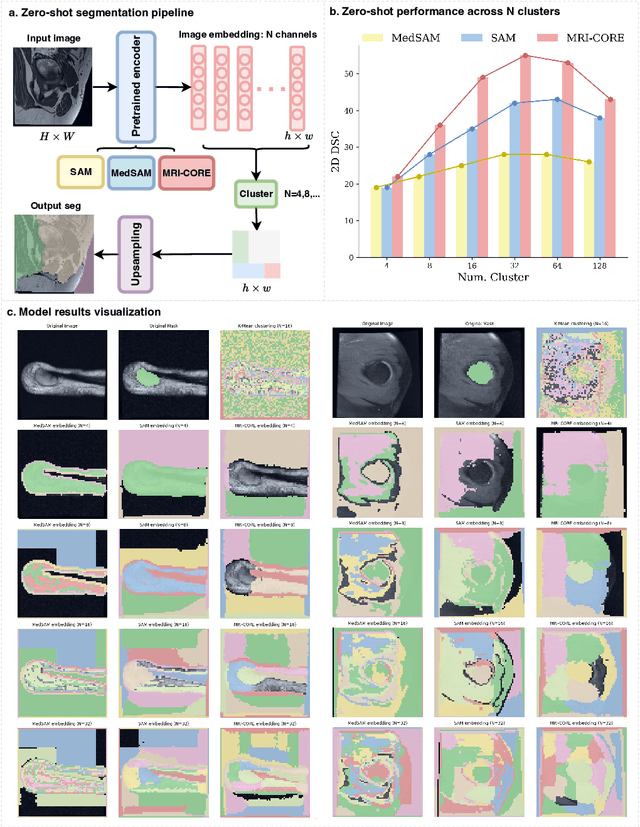
Abstract:The widespread use of Magnetic Resonance Imaging (MRI) and the rise of deep learning have enabled the development of powerful predictive models for a wide range of diagnostic tasks in MRI, such as image classification or object segmentation. However, training models for specific new tasks often requires large amounts of labeled data, which is difficult to obtain due to high annotation costs and data privacy concerns. To circumvent this issue, we introduce MRI-CORE (MRI COmprehensive Representation Encoder), a vision foundation model pre-trained using more than 6 million slices from over 110,000 MRI volumes across 18 main body locations. Experiments on five diverse object segmentation tasks in MRI demonstrate that MRI-CORE can significantly improve segmentation performance in realistic scenarios with limited labeled data availability, achieving an average gain of 6.97% 3D Dice Coefficient using only 10 annotated slices per task. We further demonstrate new model capabilities in MRI such as classification of image properties including body location, sequence type and institution, and zero-shot segmentation. These results highlight the value of MRI-CORE as a generalist vision foundation model for MRI, potentially lowering the data annotation resource barriers for many applications.
GuidedMorph: Two-Stage Deformable Registration for Breast MRI
May 19, 2025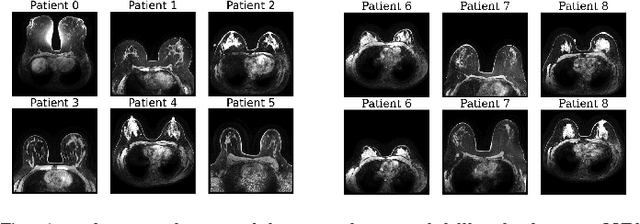
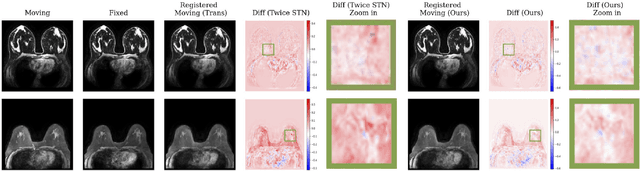
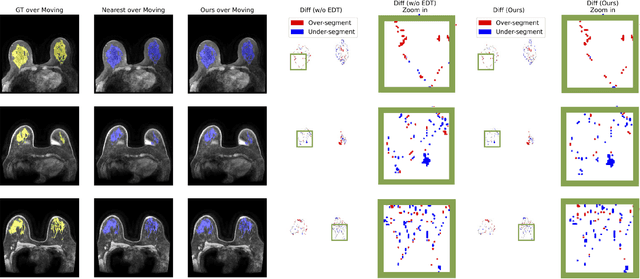
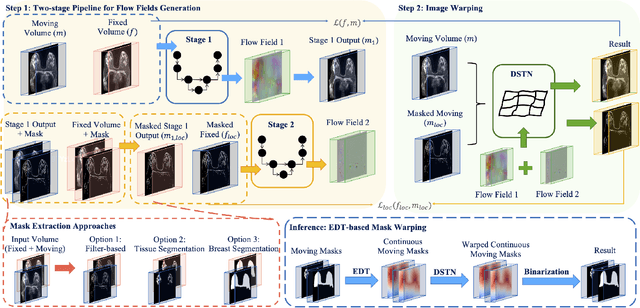
Abstract:Accurately registering breast MR images from different time points enables the alignment of anatomical structures and tracking of tumor progression, supporting more effective breast cancer detection, diagnosis, and treatment planning. However, the complexity of dense tissue and its highly non-rigid nature pose challenges for conventional registration methods, which primarily focus on aligning general structures while overlooking intricate internal details. To address this, we propose \textbf{GuidedMorph}, a novel two-stage registration framework designed to better align dense tissue. In addition to a single-scale network for global structure alignment, we introduce a framework that utilizes dense tissue information to track breast movement. The learned transformation fields are fused by introducing the Dual Spatial Transformer Network (DSTN), improving overall alignment accuracy. A novel warping method based on the Euclidean distance transform (EDT) is also proposed to accurately warp the registered dense tissue and breast masks, preserving fine structural details during deformation. The framework supports paradigms that require external segmentation models and with image data only. It also operates effectively with the VoxelMorph and TransMorph backbones, offering a versatile solution for breast registration. We validate our method on ISPY2 and internal dataset, demonstrating superior performance in dense tissue, overall breast alignment, and breast structural similarity index measure (SSIM), with notable improvements by over 13.01% in dense tissue Dice, 3.13% in breast Dice, and 1.21% in breast SSIM compared to the best learning-based baseline.
Accelerating Volumetric Medical Image Annotation via Short-Long Memory SAM 2
May 03, 2025Abstract:Manual annotation of volumetric medical images, such as magnetic resonance imaging (MRI) and computed tomography (CT), is a labor-intensive and time-consuming process. Recent advancements in foundation models for video object segmentation, such as Segment Anything Model 2 (SAM 2), offer a potential opportunity to significantly speed up the annotation process by manually annotating one or a few slices and then propagating target masks across the entire volume. However, the performance of SAM 2 in this context varies. Our experiments show that relying on a single memory bank and attention module is prone to error propagation, particularly at boundary regions where the target is present in the previous slice but absent in the current one. To address this problem, we propose Short-Long Memory SAM 2 (SLM-SAM 2), a novel architecture that integrates distinct short-term and long-term memory banks with separate attention modules to improve segmentation accuracy. We evaluate SLM-SAM 2 on three public datasets covering organs, bones, and muscles across MRI and CT modalities. We show that the proposed method markedly outperforms the default SAM 2, achieving average Dice Similarity Coefficient improvement of 0.14 and 0.11 in the scenarios when 5 volumes and 1 volume are available for the initial adaptation, respectively. SLM-SAM 2 also exhibits stronger resistance to over-propagation, making a notable step toward more accurate automated annotation of medical images for segmentation model development.
Temporal Gaussian Copula For Clinical Multivariate Time Series Data Imputation
Apr 03, 2025Abstract:The imputation of the Multivariate time series (MTS) is particularly challenging since the MTS typically contains irregular patterns of missing values due to various factors such as instrument failures, interference from irrelevant data, and privacy regulations. Existing statistical methods and deep learning methods have shown promising results in time series imputation. In this paper, we propose a Temporal Gaussian Copula Model (TGC) for three-order MTS imputation. The key idea is to leverage the Gaussian Copula to explore the cross-variable and temporal relationships based on the latent Gaussian representation. Subsequently, we employ an Expectation-Maximization (EM) algorithm to improve robustness in managing data with varying missing rates. Comprehensive experiments were conducted on three real-world MTS datasets. The results demonstrate that our TGC substantially outperforms the state-of-the-art imputation methods. Additionally, the TGC model exhibits stronger robustness to the varying missing ratios in the test dataset. Our code is available at https://github.com/MVL-Lab/TGC-MTS.
RaD: A Metric for Medical Image Distribution Comparison in Out-of-Domain Detection and Other Applications
Dec 02, 2024Abstract:Determining whether two sets of images belong to the same or different domain is a crucial task in modern medical image analysis and deep learning, where domain shift is a common problem that commonly results in decreased model performance. This determination is also important to evaluate the output quality of generative models, e.g., image-to-image translation models used to mitigate domain shift. Current metrics for this either rely on the (potentially biased) choice of some downstream task such as segmentation, or adopt task-independent perceptual metrics (e.g., FID) from natural imaging which insufficiently capture anatomical consistency and realism in medical images. We introduce a new perceptual metric tailored for medical images: Radiomic Feature Distance (RaD), which utilizes standardized, clinically meaningful and interpretable image features. We show that RaD is superior to other metrics for out-of-domain (OOD) detection in a variety of experiments. Furthermore, RaD outperforms previous perceptual metrics (FID, KID, etc.) for image-to-image translation by correlating more strongly with downstream task performance as well as anatomical consistency and realism, and shows similar utility for evaluating unconditional image generation. RaD also offers additional benefits such as interpretability, as well as stability and computational efficiency at low sample sizes. Our results are supported by broad experiments spanning four multi-domain medical image datasets, nine downstream tasks, six image translation models, and other factors, highlighting the broad potential of RaD for medical image analysis.
SAM & SAM 2 in 3D Slicer: SegmentWithSAM Extension for Annotating Medical Images
Aug 27, 2024Abstract:Creating annotations for 3D medical data is time-consuming and often requires highly specialized expertise. Various tools have been implemented to aid this process. Segment Anything Model 2 (SAM 2) offers a general-purpose prompt-based segmentation algorithm designed to annotate videos. In this paper, we adapt this model to the annotation of 3D medical images and offer our implementation in the form of an extension to the popular annotation software: 3D Slicer. Our extension allows users to place point prompts on 2D slices to generate annotation masks and propagate these annotations across entire volumes in either single-directional or bi-directional manners. Our code is publicly available on https://github.com/mazurowski-lab/SlicerSegmentWithSAM and can be easily installed directly from the Extension Manager of 3D Slicer as well.
Rethinking Perceptual Metrics for Medical Image Translation
Apr 10, 2024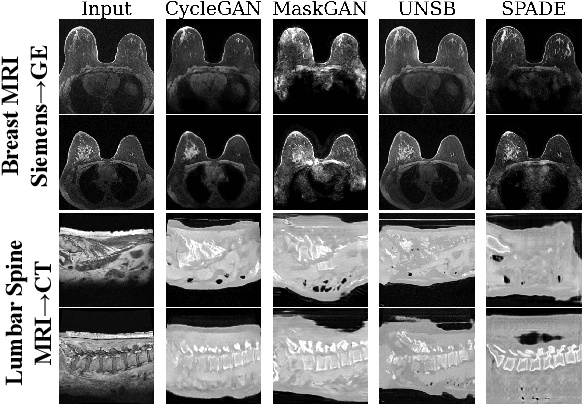
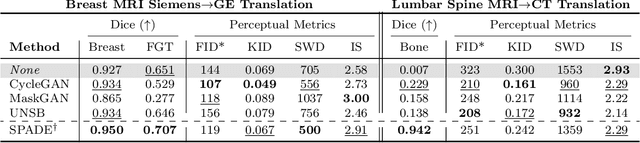
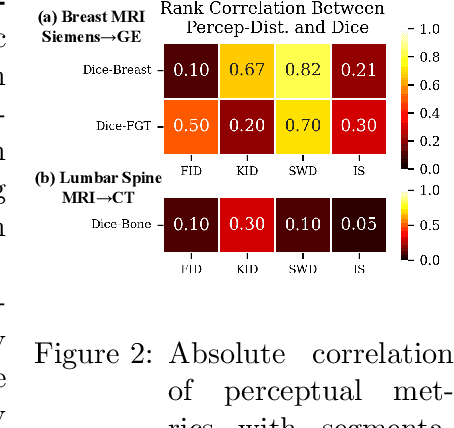
Abstract:Modern medical image translation methods use generative models for tasks such as the conversion of CT images to MRI. Evaluating these methods typically relies on some chosen downstream task in the target domain, such as segmentation. On the other hand, task-agnostic metrics are attractive, such as the network feature-based perceptual metrics (e.g., FID) that are common to image translation in general computer vision. In this paper, we investigate evaluation metrics for medical image translation on two medical image translation tasks (GE breast MRI to Siemens breast MRI and lumbar spine MRI to CT), tested on various state-of-the-art translation methods. We show that perceptual metrics do not generally correlate with segmentation metrics due to them extending poorly to the anatomical constraints of this sub-field, with FID being especially inconsistent. However, we find that the lesser-used pixel-level SWD metric may be useful for subtle intra-modality translation. Our results demonstrate the need for further research into helpful metrics for medical image translation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge