Brian Guo
The Impact of Scanner Domain Shift on Deep Learning Performance in Medical Imaging: an Experimental Study
Sep 06, 2024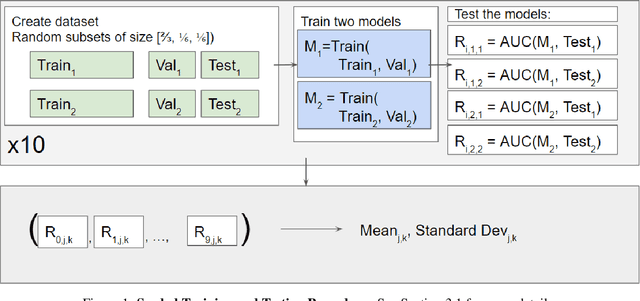
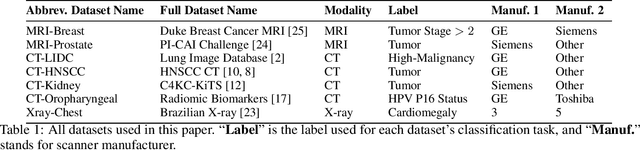
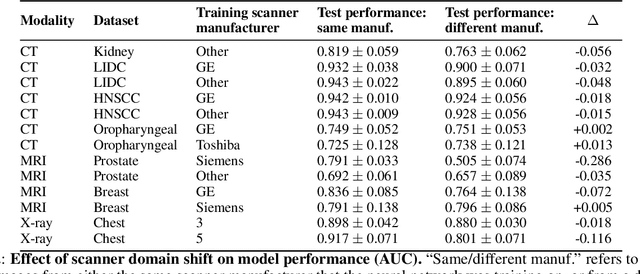
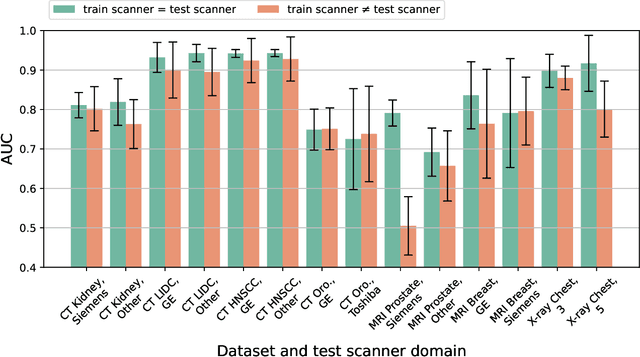
Abstract:Purpose: Medical images acquired using different scanners and protocols can differ substantially in their appearance. This phenomenon, scanner domain shift, can result in a drop in the performance of deep neural networks which are trained on data acquired by one scanner and tested on another. This significant practical issue is well-acknowledged, however, no systematic study of the issue is available across different modalities and diagnostic tasks. Materials and Methods: In this paper, we present a broad experimental study evaluating the impact of scanner domain shift on convolutional neural network performance for different automated diagnostic tasks. We evaluate this phenomenon in common radiological modalities, including X-ray, CT, and MRI. Results: We find that network performance on data from a different scanner is almost always worse than on same-scanner data, and we quantify the degree of performance drop across different datasets. Notably, we find that this drop is most severe for MRI, moderate for X-ray, and quite small for CT, on average, which we attribute to the standardized nature of CT acquisition systems which is not present in MRI or X-ray. We also study how injecting varying amounts of target domain data into the training set, as well as adding noise to the training data, helps with generalization. Conclusion: Our results provide extensive experimental evidence and quantification of the extent of performance drop caused by scanner domain shift in deep learning across different modalities, with the goal of guiding the future development of robust deep learning models for medical image analysis.
SegmentAnyBone: A Universal Model that Segments Any Bone at Any Location on MRI
Jan 23, 2024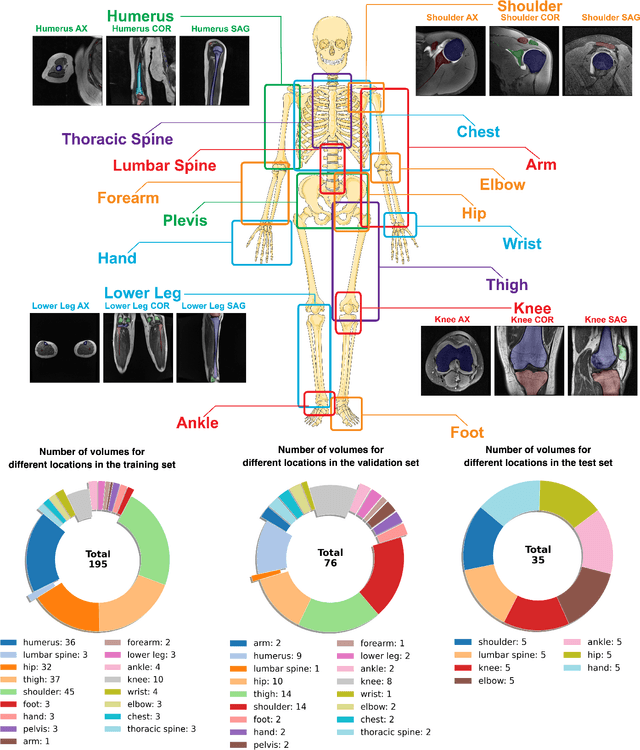
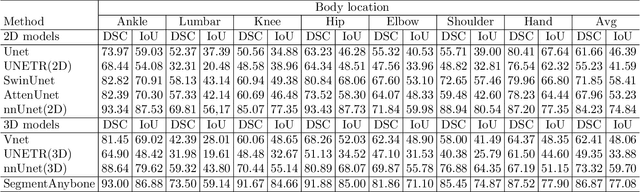
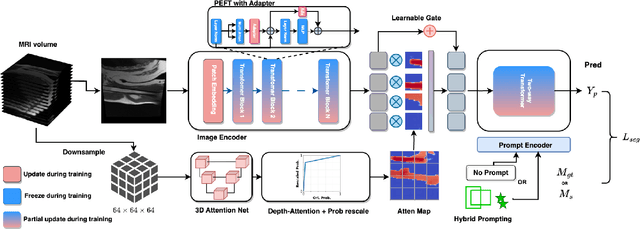

Abstract:Magnetic Resonance Imaging (MRI) is pivotal in radiology, offering non-invasive and high-quality insights into the human body. Precise segmentation of MRIs into different organs and tissues would be highly beneficial since it would allow for a higher level of understanding of the image content and enable important measurements, which are essential for accurate diagnosis and effective treatment planning. Specifically, segmenting bones in MRI would allow for more quantitative assessments of musculoskeletal conditions, while such assessments are largely absent in current radiological practice. The difficulty of bone MRI segmentation is illustrated by the fact that limited algorithms are publicly available for use, and those contained in the literature typically address a specific anatomic area. In our study, we propose a versatile, publicly available deep-learning model for bone segmentation in MRI across multiple standard MRI locations. The proposed model can operate in two modes: fully automated segmentation and prompt-based segmentation. Our contributions include (1) collecting and annotating a new MRI dataset across various MRI protocols, encompassing over 300 annotated volumes and 8485 annotated slices across diverse anatomic regions; (2) investigating several standard network architectures and strategies for automated segmentation; (3) introducing SegmentAnyBone, an innovative foundational model-based approach that extends Segment Anything Model (SAM); (4) comparative analysis of our algorithm and previous approaches; and (5) generalization analysis of our algorithm across different anatomical locations and MRI sequences, as well as an external dataset. We publicly release our model at https://github.com/mazurowski-lab/SegmentAnyBone.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge