Binxu Li
Integrating Anatomical Priors into a Causal Diffusion Model
Sep 10, 2025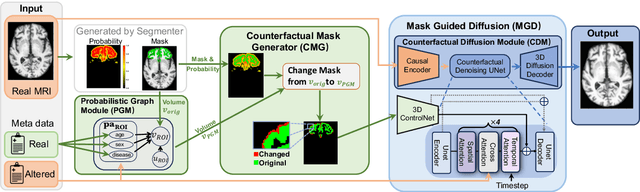
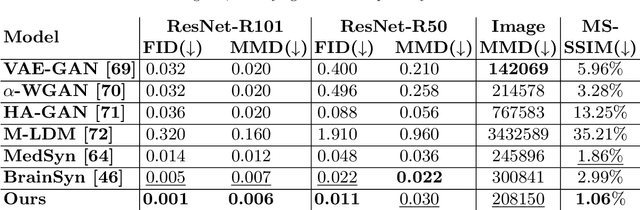
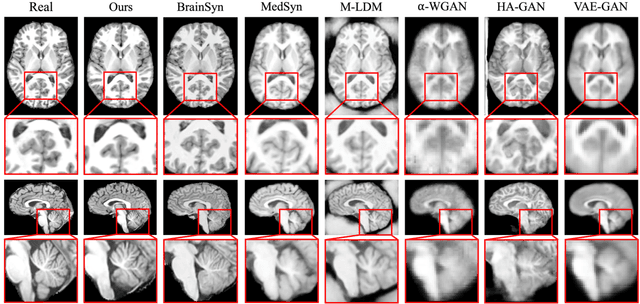

Abstract:3D brain MRI studies often examine subtle morphometric differences between cohorts that are hard to detect visually. Given the high cost of MRI acquisition, these studies could greatly benefit from image syntheses, particularly counterfactual image generation, as seen in other domains, such as computer vision. However, counterfactual models struggle to produce anatomically plausible MRIs due to the lack of explicit inductive biases to preserve fine-grained anatomical details. This shortcoming arises from the training of the models aiming to optimize for the overall appearance of the images (e.g., via cross-entropy) rather than preserving subtle, yet medically relevant, local variations across subjects. To preserve subtle variations, we propose to explicitly integrate anatomical constraints on a voxel-level as prior into a generative diffusion framework. Called Probabilistic Causal Graph Model (PCGM), the approach captures anatomical constraints via a probabilistic graph module and translates those constraints into spatial binary masks of regions where subtle variations occur. The masks (encoded by a 3D extension of ControlNet) constrain a novel counterfactual denoising UNet, whose encodings are then transferred into high-quality brain MRIs via our 3D diffusion decoder. Extensive experiments on multiple datasets demonstrate that PCGM generates structural brain MRIs of higher quality than several baseline approaches. Furthermore, we show for the first time that brain measurements extracted from counterfactuals (generated by PCGM) replicate the subtle effects of a disease on cortical brain regions previously reported in the neuroscience literature. This achievement is an important milestone in the use of synthetic MRIs in studies investigating subtle morphological differences.
Divergence Minimization Preference Optimization for Diffusion Model Alignment
Jul 10, 2025Abstract:Diffusion models have achieved remarkable success in generating realistic and versatile images from text prompts. Inspired by the recent advancements of language models, there is an increasing interest in further improving the models by aligning with human preferences. However, we investigate alignment from a divergence minimization perspective and reveal that existing preference optimization methods are typically trapped in suboptimal mean-seeking optimization. In this paper, we introduce Divergence Minimization Preference Optimization (DMPO), a novel and principled method for aligning diffusion models by minimizing reverse KL divergence, which asymptotically enjoys the same optimization direction as original RL. We provide rigorous analysis to justify the effectiveness of DMPO and conduct comprehensive experiments to validate its empirical strength across both human evaluations and automatic metrics. Our extensive results show that diffusion models fine-tuned with DMPO can consistently outperform or match existing techniques, specifically outperforming all existing diffusion alignment baselines by at least 64.6% in PickScore across all evaluation datasets, demonstrating the method's superiority in aligning generative behavior with desired outputs. Overall, DMPO unlocks a robust and elegant pathway for preference alignment, bridging principled theory with practical performance in diffusion models.
Efficient Temporal Consistency in Diffusion-Based Video Editing with Adaptor Modules: A Theoretical Framework
Apr 22, 2025Abstract:Adapter-based methods are commonly used to enhance model performance with minimal additional complexity, especially in video editing tasks that require frame-to-frame consistency. By inserting small, learnable modules into pretrained diffusion models, these adapters can maintain temporal coherence without extensive retraining. Approaches that incorporate prompt learning with both shared and frame-specific tokens are particularly effective in preserving continuity across frames at low training cost. In this work, we want to provide a general theoretical framework for adapters that maintain frame consistency in DDIM-based models under a temporal consistency loss. First, we prove that the temporal consistency objective is differentiable under bounded feature norms, and we establish a Lipschitz bound on its gradient. Second, we show that gradient descent on this objective decreases the loss monotonically and converges to a local minimum if the learning rate is within an appropriate range. Finally, we analyze the stability of modules in the DDIM inversion procedure, showing that the associated error remains controlled. These theoretical findings will reinforce the reliability of diffusion-based video editing methods that rely on adapter strategies and provide theoretical insights in video generation tasks.
Enhancing Low-Cost Video Editing with Lightweight Adaptors and Temporal-Aware Inversion
Jan 08, 2025



Abstract:Recent advancements in text-to-image (T2I) generation using diffusion models have enabled cost-effective video-editing applications by leveraging pre-trained models, eliminating the need for resource-intensive training. However, the frame-independence of T2I generation often results in poor temporal consistency. Existing methods address this issue through temporal layer fine-tuning or inference-based temporal propagation, but these approaches suffer from high training costs or limited temporal coherence. To address these challenges, we propose a General and Efficient Adapter (GE-Adapter) that integrates temporal-spatial and semantic consistency with Baliteral DDIM inversion. This framework introduces three key components: (1) Frame-based Temporal Consistency Blocks (FTC Blocks) to capture frame-specific features and enforce smooth inter-frame transitions via temporally-aware loss functions; (2) Channel-dependent Spatial Consistency Blocks (SCD Blocks) employing bilateral filters to enhance spatial coherence by reducing noise and artifacts; and (3) Token-based Semantic Consistency Module (TSC Module) to maintain semantic alignment using shared prompt tokens and frame-specific tokens. Our method significantly improves perceptual quality, text-image alignment, and temporal coherence, as demonstrated on the MSR-VTT dataset. Additionally, it achieves enhanced fidelity and frame-to-frame coherence, offering a practical solution for T2V editing.
Evaluating the Quality of Brain MRI Generators
Sep 13, 2024Abstract:Deep learning models generating structural brain MRIs have the potential to significantly accelerate discovery of neuroscience studies. However, their use has been limited in part by the way their quality is evaluated. Most evaluations of generative models focus on metrics originally designed for natural images (such as structural similarity index and Frechet inception distance). As we show in a comparison of 6 state-of-the-art generative models trained and tested on over 3000 MRIs, these metrics are sensitive to the experimental setup and inadequately assess how well brain MRIs capture macrostructural properties of brain regions (i.e., anatomical plausibility). This shortcoming of the metrics results in inconclusive findings even when qualitative differences between the outputs of models are evident. We therefore propose a framework for evaluating models generating brain MRIs, which requires uniform processing of the real MRIs, standardizing the implementation of the models, and automatically segmenting the MRIs generated by the models. The segmentations are used for quantifying the plausibility of anatomy displayed in the MRIs. To ensure meaningful quantification, it is crucial that the segmentations are highly reliable. Our framework rigorously checks this reliability, a step often overlooked by prior work. Only 3 of the 6 generative models produced MRIs, of which at least 95% had highly reliable segmentations. More importantly, the assessment of each model by our framework is in line with qualitative assessments, reinforcing the validity of our approach.
MMedAgent: Learning to Use Medical Tools with Multi-modal Agent
Jul 02, 2024



Abstract:Multi-Modal Large Language Models (MLLMs), despite being successful, exhibit limited generality and often fall short when compared to specialized models. Recently, LLM-based agents have been developed to address these challenges by selecting appropriate specialized models as tools based on user inputs. However, such advancements have not been extensively explored within the medical domain. To bridge this gap, this paper introduces the first agent explicitly designed for the medical field, named \textbf{M}ulti-modal \textbf{Med}ical \textbf{Agent} (MMedAgent). We curate an instruction-tuning dataset comprising six medical tools solving seven tasks, enabling the agent to choose the most suitable tools for a given task. Comprehensive experiments demonstrate that MMedAgent achieves superior performance across a variety of medical tasks compared to state-of-the-art open-source methods and even the closed-source model, GPT-4o. Furthermore, MMedAgent exhibits efficiency in updating and integrating new medical tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge