Jinwei Zhang
MSRepaint: Multiple Sclerosis Repaint with Conditional Denoising Diffusion Implicit Model for Bidirectional Lesion Filling and Synthesis
Oct 02, 2025Abstract:In multiple sclerosis, lesions interfere with automated magnetic resonance imaging analyses such as brain parcellation and deformable registration, while lesion segmentation models are hindered by the limited availability of annotated training data. To address both issues, we propose MSRepaint, a unified diffusion-based generative model for bidirectional lesion filling and synthesis that restores anatomical continuity for downstream analyses and augments segmentation through realistic data generation. MSRepaint conditions on spatial lesion masks for voxel-level control, incorporates contrast dropout to handle missing inputs, integrates a repainting mechanism to preserve surrounding anatomy during lesion filling and synthesis, and employs a multi-view DDIM inversion and fusion pipeline for 3D consistency with fast inference. Extensive evaluations demonstrate the effectiveness of MSRepaint across multiple tasks. For lesion filling, we evaluate both the accuracy within the filled regions and the impact on downstream tasks including brain parcellation and deformable registration. MSRepaint outperforms the traditional lesion filling methods FSL and NiftySeg, and achieves accuracy on par with FastSurfer-LIT, a recent diffusion model-based inpainting method, while offering over 20 times faster inference. For lesion synthesis, state-of-the-art MS lesion segmentation models trained on MSRepaint-synthesized data outperform those trained on CarveMix-synthesized data or real ISBI challenge training data across multiple benchmarks, including the MICCAI 2016 and UMCL datasets. Additionally, we demonstrate that MSRepaint's unified bidirectional filling and synthesis capability, with full spatial control over lesion appearance, enables high-fidelity simulation of lesion evolution in longitudinal MS progression.
UNISELF: A Unified Network with Instance Normalization and Self-Ensembled Lesion Fusion for Multiple Sclerosis Lesion Segmentation
Aug 06, 2025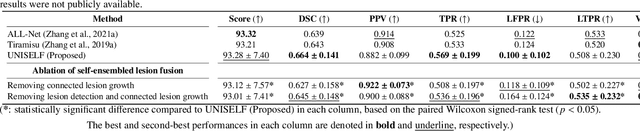
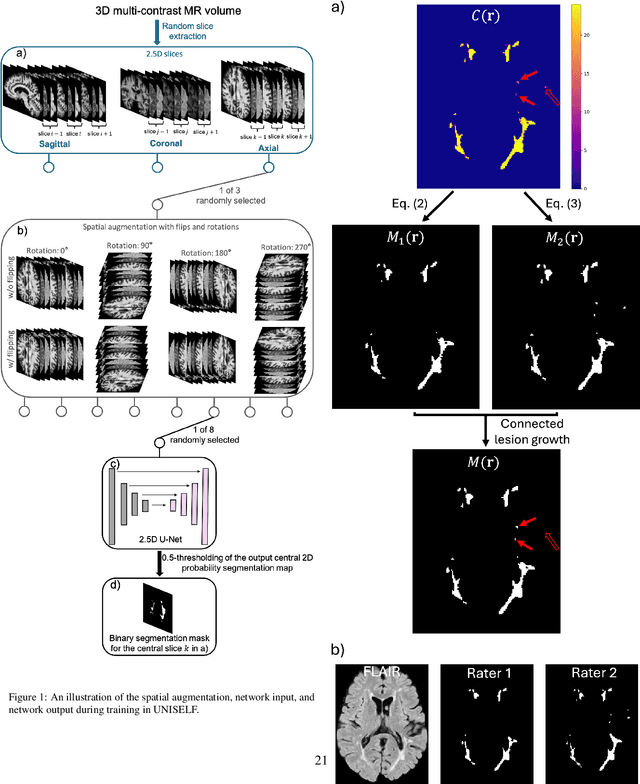
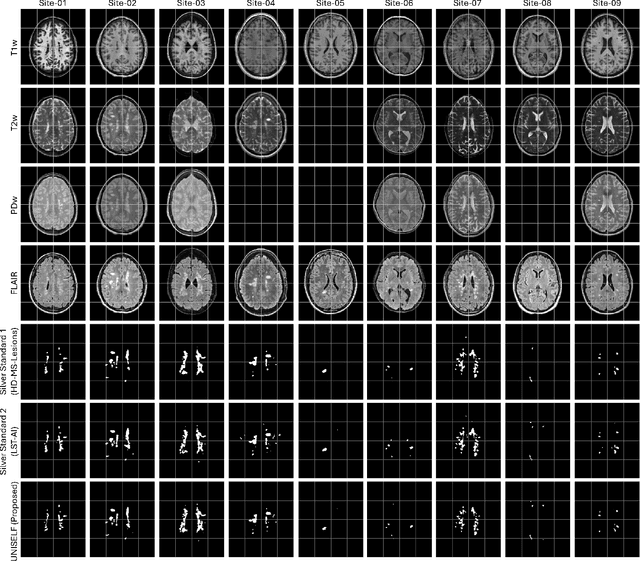

Abstract:Automated segmentation of multiple sclerosis (MS) lesions using multicontrast magnetic resonance (MR) images improves efficiency and reproducibility compared to manual delineation, with deep learning (DL) methods achieving state-of-the-art performance. However, these DL-based methods have yet to simultaneously optimize in-domain accuracy and out-of-domain generalization when trained on a single source with limited data, or their performance has been unsatisfactory. To fill this gap, we propose a method called UNISELF, which achieves high accuracy within a single training domain while demonstrating strong generalizability across multiple out-of-domain test datasets. UNISELF employs a novel test-time self-ensembled lesion fusion to improve segmentation accuracy, and leverages test-time instance normalization (TTIN) of latent features to address domain shifts and missing input contrasts. Trained on the ISBI 2015 longitudinal MS segmentation challenge training dataset, UNISELF ranks among the best-performing methods on the challenge test dataset. Additionally, UNISELF outperforms all benchmark methods trained on the same ISBI training data across diverse out-of-domain test datasets with domain shifts and missing contrasts, including the public MICCAI 2016 and UMCL datasets, as well as a private multisite dataset. These test datasets exhibit domain shifts and/or missing contrasts caused by variations in acquisition protocols, scanner types, and imaging artifacts arising from imperfect acquisition. Our code is available at https://github.com/uponacceptance.
ECLARE: Efficient cross-planar learning for anisotropic resolution enhancement
Mar 14, 2025Abstract:In clinical imaging, magnetic resonance (MR) image volumes are often acquired as stacks of 2D slices, permitting decreased scan times, improved signal-to-noise ratio, and image contrasts unique to 2D MR pulse sequences. While this is sufficient for clinical evaluation, automated algorithms designed for 3D analysis perform sub-optimally on 2D-acquired scans, especially those with thick slices and gaps between slices. Super-resolution (SR) methods aim to address this problem, but previous methods do not address all of the following: slice profile shape estimation, slice gap, domain shift, and non-integer / arbitrary upsampling factors. In this paper, we propose ECLARE (Efficient Cross-planar Learning for Anisotropic Resolution Enhancement), a self-SR method that addresses each of these factors. ECLARE estimates the slice profile from the 2D-acquired multi-slice MR volume, trains a network to learn the mapping from low-resolution to high-resolution in-plane patches from the same volume, and performs SR with anti-aliasing. We compared ECLARE to cubic B-spline interpolation, SMORE, and other contemporary SR methods. We used realistic and representative simulations so that quantitative performance against a ground truth could be computed, and ECLARE outperformed all other methods in both signal recovery and downstream tasks. On real data for which there is no ground truth, ECLARE demonstrated qualitative superiority over other methods as well. Importantly, as ECLARE does not use external training data it cannot suffer from domain shift between training and testing. Our code is open-source and available at https://www.github.com/sremedios/eclare.
Implicit neural representation for free-breathing MR fingerprinting (INR-MRF): co-registered 3D whole-liver water T1, water T2, proton density fat fraction, and R2* mapping
Oct 19, 2024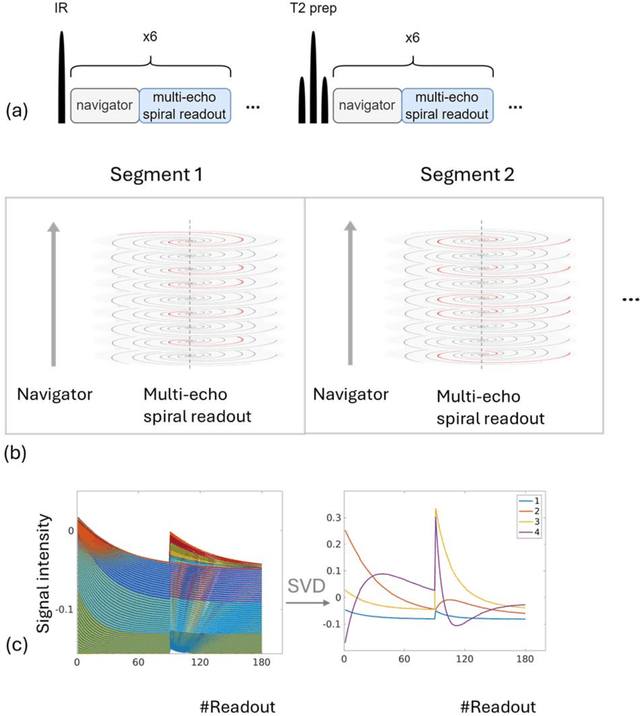

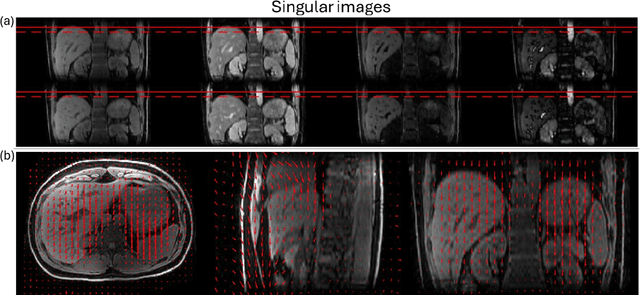
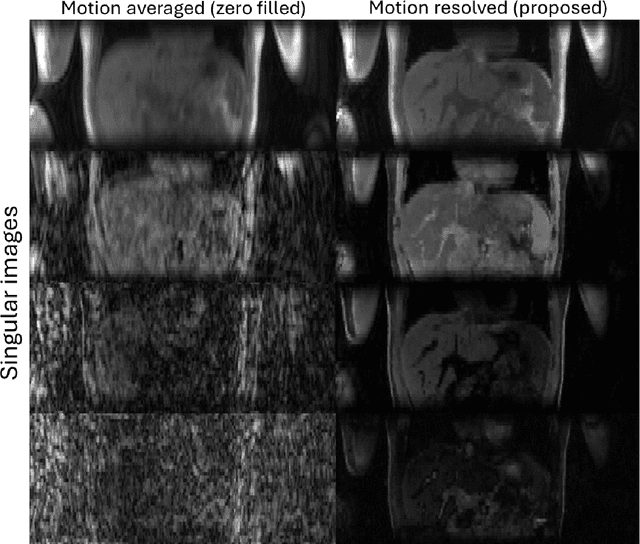
Abstract:Purpose: To develop an MRI technique for free-breathing 3D whole-liver quantification of water T1, water T2, proton density fat fraction (PDFF), R2*. Methods: An Eight-echo spoiled gradient echo pulse sequence with spiral readout was developed by interleaving inversion recovery and T2 magnetization preparation. We propose a neural network based on a 4D and a 3D implicit neural representation (INR) which simultaneously learns the motion deformation fields and the static reference frame MRI subspace images respectively. Water and fat singular images were separated during network training, with no need of performing retrospective water-fat separation. T1, T2, R2* and proton density fat fraction (PDFF) produced by the proposed method were validated in vivo on 10 healthy subjects, using quantitative maps generated from conventional scans as reference. Results: Our results showed minimal bias and narrow 95% limits of agreement on T1, T2, R2* and PDFF values in the liver compared to conventional breath-holding scans. Conclusions: INR-MRF enabled co-registered 3D whole liver T1, T2, R2* and PDFF mapping in a single free-breathing scan.
Unique MS Lesion Identification from MRI
Oct 12, 2024



Abstract:Unique identification of multiple sclerosis (MS) white matter lesions (WMLs) is important to help characterize MS progression. WMLs are routinely identified from magnetic resonance images (MRIs) but the resultant total lesion load does not correlate well with EDSS; whereas mean unique lesion volume has been shown to correlate with EDSS. Our approach builds on prior work by incorporating Hessian matrix computation from lesion probability maps before using the random walker algorithm to estimate the volume of each unique lesion. Synthetic images demonstrate our ability to accurately count the number of lesions present. The takeaways, are: 1) that our method correctly identifies all lesions including many that are missed by previous methods; 2) we can better separate confluent lesions; and 3) we can accurately capture the total volume of WMLs in a given probability map. This work will allow new more meaningful statistics to be computed from WMLs in brain MRIs
Bi-Directional MS Lesion Filling and Synthesis Using Denoising Diffusion Implicit Model-based Lesion Repainting
Oct 07, 2024Abstract:Automatic magnetic resonance (MR) image processing pipelines are widely used to study people with multiple sclerosis (PwMS), encompassing tasks such as lesion segmentation and brain parcellation. However, the presence of lesion often complicates these analysis, particularly in brain parcellation. Lesion filling is commonly used to mitigate this issue, but existing lesion filling algorithms often fall short in accurately reconstructing realistic lesion-free images, which are vital for consistent downstream analysis. Additionally, the performance of lesion segmentation algorithms is often limited by insufficient data with lesion delineation as training labels. In this paper, we propose a novel approach leveraging Denoising Diffusion Implicit Models (DDIMs) for both MS lesion filling and synthesis based on image inpainting. Our modified DDIM architecture, once trained, enables both MS lesion filing and synthesis. Specifically, it can generate lesion-free T1-weighted or FLAIR images from those containing lesions; Or it can add lesions to T1-weighted or FLAIR images of healthy subjects. The former is essential for downstream analyses that require lesion-free images, while the latter is valuable for augmenting training datasets for lesion segmentation tasks. We validate our approach through initial experiments in this paper and demonstrate promising results in both lesion filling and synthesis, paving the way for future work.
MRI quantification of liver fibrosis using diamagnetic susceptibility: An ex-vivo feasibility study
Oct 04, 2024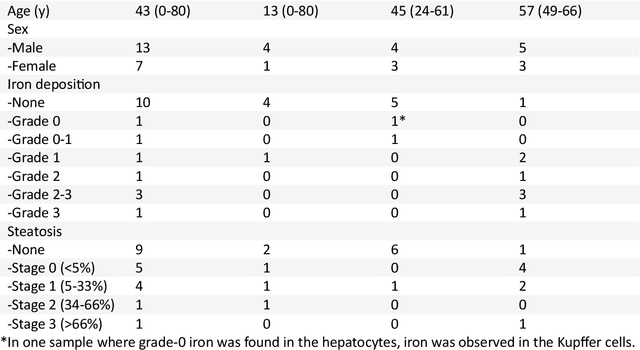
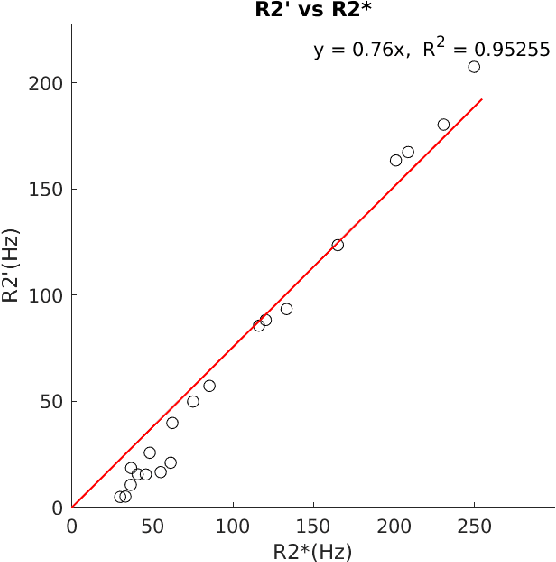
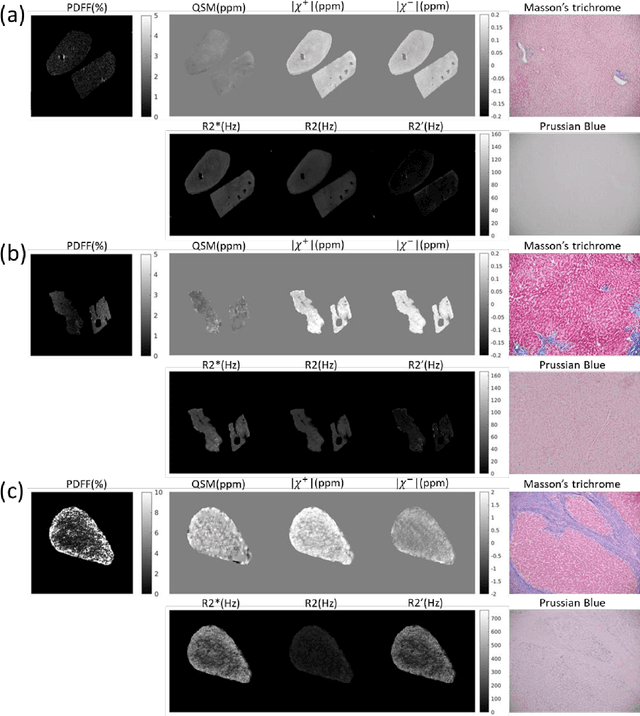
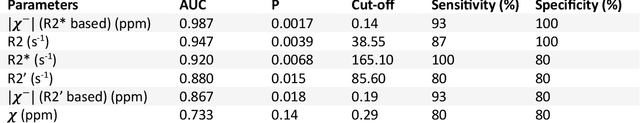
Abstract:In chronic liver disease, liver fibrosis develops as excessive deposition of extracellular matrix macromolecules, predominantly collagens, progressively form fibrous scars that disrupt the hepatic architecture, and fibrosis, iron, and fat are interrelated. Fibrosis is the best predictor of morbidity and mortality in chronic liver disease but liver biopsy, the reference method for diagnosis and staging, is invasive and limited by sampling and interobserver variability and risks of complications. The overall objective of this study was to develop a new non-invasive method to quantify fibrosis using diamagnetic susceptibility sources with histology validation in ex vivo liver explants.
STimage-1K4M: A histopathology image-gene expression dataset for spatial transcriptomics
Jun 10, 2024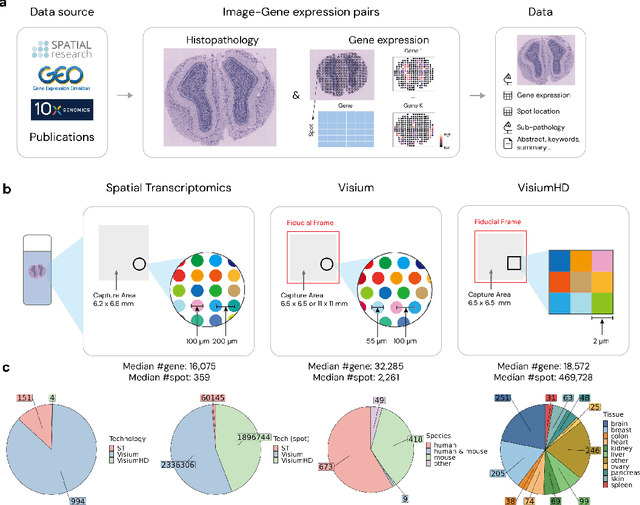
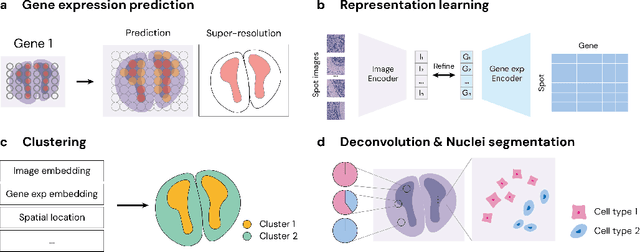
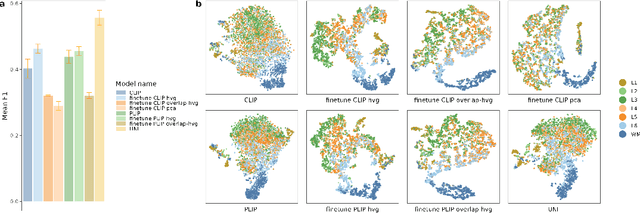
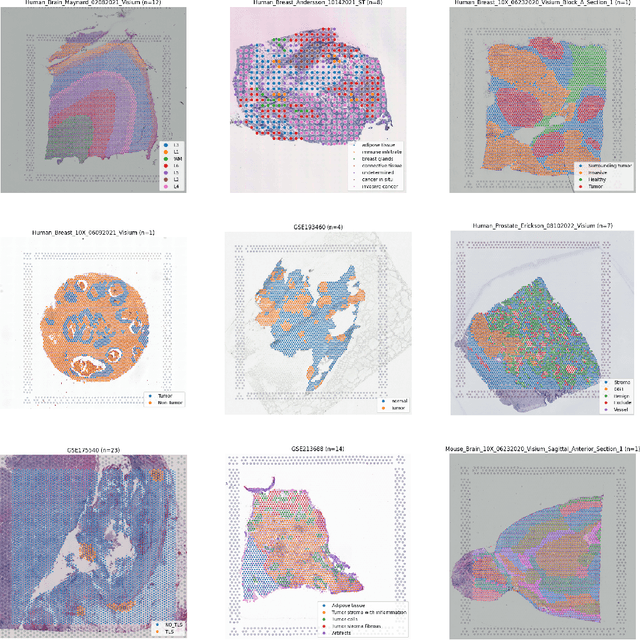
Abstract:Recent advances in multi-modal algorithms have driven and been driven by the increasing availability of large image-text datasets, leading to significant strides in various fields, including computational pathology. However, in most existing medical image-text datasets, the text typically provides high-level summaries that may not sufficiently describe sub-tile regions within a large pathology image. For example, an image might cover an extensive tissue area containing cancerous and healthy regions, but the accompanying text might only specify that this image is a cancer slide, lacking the nuanced details needed for in-depth analysis. In this study, we introduce STimage-1K4M, a novel dataset designed to bridge this gap by providing genomic features for sub-tile images. STimage-1K4M contains 1,149 images derived from spatial transcriptomics data, which captures gene expression information at the level of individual spatial spots within a pathology image. Specifically, each image in the dataset is broken down into smaller sub-image tiles, with each tile paired with 15,000-30,000 dimensional gene expressions. With 4,293,195 pairs of sub-tile images and gene expressions, STimage-1K4M offers unprecedented granularity, paving the way for a wide range of advanced research in multi-modal data analysis an innovative applications in computational pathology, and beyond.
Deep learning improved autofocus for motion artifact reduction and its application in quantitative susceptibility mapping
May 26, 2024Abstract:Purpose: To develop a pipeline for motion artifact correction in mGRE and quantitative susceptibility mapping (QSM). Methods: Deep learning is integrated with autofocus to improve motion artifact suppression, which is applied QSM of patients with Parkinson's disease (PD). The estimation of affine motion parameters in the autofocus method depends on signal-to-noise ratio and lacks accuracy when data sampling occurs outside the k-space center. A deep learning strategy is employed to remove the residual motion artifacts in autofocus. Results: Results obtained in simulated brain data (n =15) with reference truth show that the proposed autofocus deep learning method significantly improves the image quality of mGRE and QSM (p = 0.001 for SSIM, p < 0.0001 for PSNR and RMSE). Results from 10 PD patients with real motion artifacts in QSM have also been corrected using the proposed method and sent to an experienced radiologist for image quality evaluation, and the average image quality score has increased (p=0.0039). Conclusions: The proposed method enables substantial suppression of motion artifacts in mGRE and QSM.
RimSet: Quantitatively Identifying and Characterizing Chronic Active Multiple Sclerosis Lesion on Quantitative Susceptibility Maps
Dec 28, 2023Abstract:Background: Rim+ lesions in multiple sclerosis (MS), detectable via Quantitative Susceptibility Mapping (QSM), correlate with increased disability. Existing literature lacks quantitative analysis of these lesions. We introduce RimSet for quantitative identification and characterization of rim+ lesions on QSM. Methods: RimSet combines RimSeg, an unsupervised segmentation method using level-set methodology, and radiomic measurements with Local Binary Pattern texture descriptors. We validated RimSet using simulated QSM images and an in vivo dataset of 172 MS subjects with 177 rim+ and 3986 rim-lesions. Results: RimSeg achieved a 78.7% Dice score against the ground truth, with challenges in partial rim lesions. RimSet detected rim+ lesions with a partial ROC AUC of 0.808 and PR AUC of 0.737, surpassing existing methods. QSMRim-Net showed the lowest mean square error (0.85) and high correlation (0.91; 95% CI: 0.88, 0.93) with expert annotations at the subject level.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge