Samuel Remedios
Bi-Directional MS Lesion Filling and Synthesis Using Denoising Diffusion Implicit Model-based Lesion Repainting
Oct 07, 2024Abstract:Automatic magnetic resonance (MR) image processing pipelines are widely used to study people with multiple sclerosis (PwMS), encompassing tasks such as lesion segmentation and brain parcellation. However, the presence of lesion often complicates these analysis, particularly in brain parcellation. Lesion filling is commonly used to mitigate this issue, but existing lesion filling algorithms often fall short in accurately reconstructing realistic lesion-free images, which are vital for consistent downstream analysis. Additionally, the performance of lesion segmentation algorithms is often limited by insufficient data with lesion delineation as training labels. In this paper, we propose a novel approach leveraging Denoising Diffusion Implicit Models (DDIMs) for both MS lesion filling and synthesis based on image inpainting. Our modified DDIM architecture, once trained, enables both MS lesion filing and synthesis. Specifically, it can generate lesion-free T1-weighted or FLAIR images from those containing lesions; Or it can add lesions to T1-weighted or FLAIR images of healthy subjects. The former is essential for downstream analyses that require lesion-free images, while the latter is valuable for augmenting training datasets for lesion segmentation tasks. We validate our approach through initial experiments in this paper and demonstrate promising results in both lesion filling and synthesis, paving the way for future work.
MR Slice Profile Estimation by Learning to Match Internal Patch Distributions
Mar 31, 2021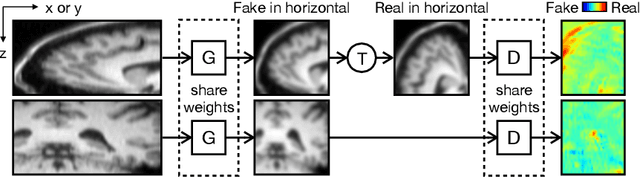
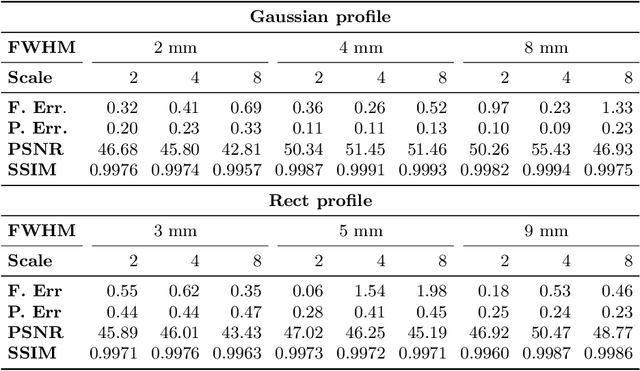
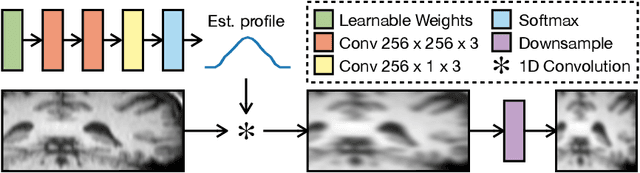
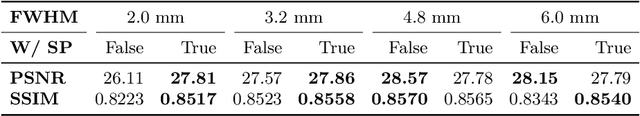
Abstract:To super-resolve the through-plane direction of a multi-slice 2D magnetic resonance (MR) image, its slice selection profile can be used as the degeneration model from high resolution (HR) to low resolution (LR) to create paired data when training a supervised algorithm. Existing super-resolution algorithms make assumptions about the slice selection profile since it is not readily known for a given image. In this work, we estimate a slice selection profile given a specific image by learning to match its internal patch distributions. Specifically, we assume that after applying the correct slice selection profile, the image patch distribution along HR in-plane directions should match the distribution along the LR through-plane direction. Therefore, we incorporate the estimation of a slice selection profile as part of learning a generator in a generative adversarial network (GAN). In this way, the slice selection profile can be learned without any external data. Our algorithm was tested using simulations from isotropic MR images, incorporated in a through-plane super-resolution algorithm to demonstrate its benefits, and also used as a tool to measure image resolution. Our code is at https://github.com/shuohan/espreso2.
Distributed deep learning for robust multi-site segmentation of CT imaging after traumatic brain injury
Mar 11, 2019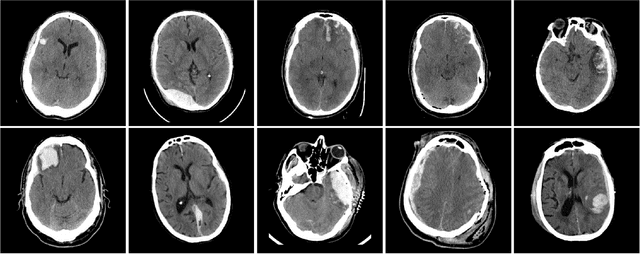

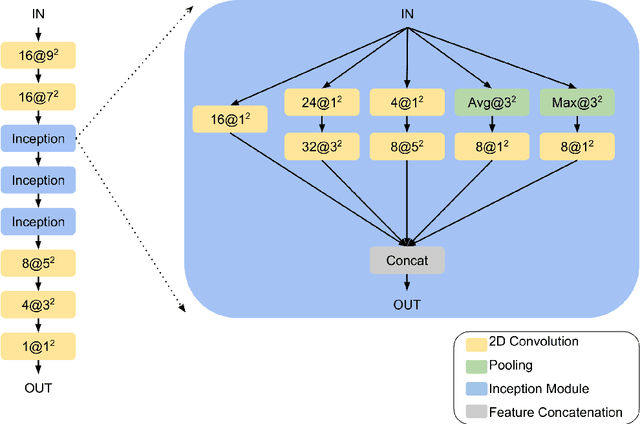
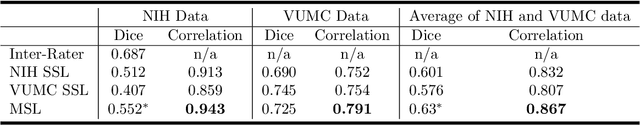
Abstract:Machine learning models are becoming commonplace in the domain of medical imaging, and with these methods comes an ever-increasing need for more data. However, to preserve patient anonymity it is frequently impractical or prohibited to transfer protected health information (PHI) between institutions. Additionally, due to the nature of some studies, there may not be a large public dataset available on which to train models. To address this conundrum, we analyze the efficacy of transferring the model itself in lieu of data between different sites. By doing so we accomplish two goals: 1) the model gains access to training on a larger dataset that it could not normally obtain and 2) the model better generalizes, having trained on data from separate locations. In this paper, we implement multi-site learning with disparate datasets from the National Institutes of Health (NIH) and Vanderbilt University Medical Center (VUMC) without compromising PHI. Three neural networks are trained to convergence on a computed tomography (CT) brain hematoma segmentation task: one only with NIH data,one only with VUMC data, and one multi-site model alternating between NIH and VUMC data. Resultant lesion masks with the multi-site model attain an average Dice similarity coefficient of 0.64 and the automatically segmented hematoma volumes correlate to those done manually with a Pearson correlation coefficient of 0.87,corresponding to an 8% and 5% improvement, respectively, over the single-site model counterparts.
Inter-Scanner Harmonization of High Angular Resolution DW-MRI using Null Space Deep Learning
Oct 09, 2018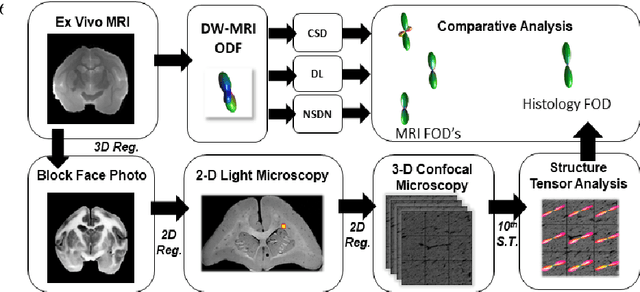
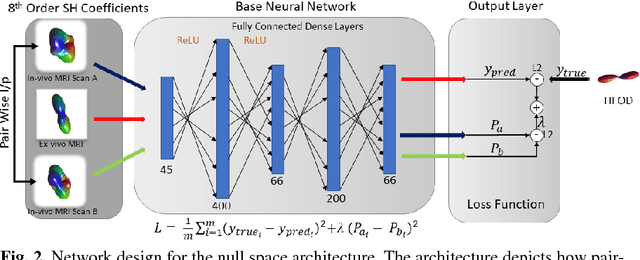

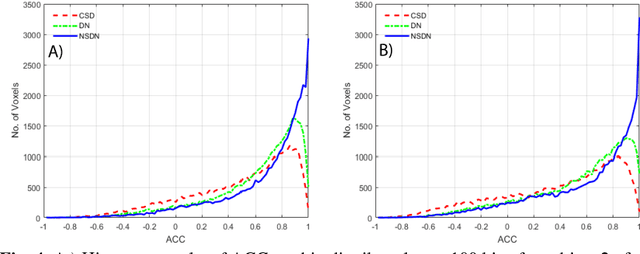
Abstract:Diffusion-weighted magnetic resonance imaging (DW-MRI) allows for non-invasive imaging of the local fiber architecture of the human brain at a millimetric scale. Multiple classical approaches have been proposed to detect both single (e.g., tensors) and multiple (e.g., constrained spherical deconvolution, CSD) fiber population orientations per voxel. However, existing techniques generally exhibit low reproducibility across MRI scanners. Herein, we propose a data-driven tech-nique using a neural network design which exploits two categories of data. First, training data were acquired on three squirrel monkey brains using ex-vivo DW-MRI and histology of the brain. Second, repeated scans of human subjects were acquired on two different scanners to augment the learning of the network pro-posed. To use these data, we propose a new network architecture, the null space deep network (NSDN), to simultaneously learn on traditional observed/truth pairs (e.g., MRI-histology voxels) along with repeated observations without a known truth (e.g., scan-rescan MRI). The NSDN was tested on twenty percent of the histology voxels that were kept completely blind to the network. NSDN significantly improved absolute performance relative to histology by 3.87% over CSD and 1.42% over a recently proposed deep neural network approach. More-over, it improved reproducibility on the paired data by 21.19% over CSD and 10.09% over a recently proposed deep approach. Finally, NSDN improved gen-eralizability of the model to a third in vivo human scanner (which was not used in training) by 16.08% over CSD and 10.41% over a recently proposed deep learn-ing approach. This work suggests that data-driven approaches for local fiber re-construction are more reproducible, informative and precise and offers a novel, practical method for determining these models.
Classifying magnetic resonance image modalities with convolutional neural networks
Apr 17, 2018Abstract:Magnetic Resonance (MR) imaging allows the acquisition of images with different contrast properties depending on the acquisition protocol and the magnetic properties of tissues. Many MR brain image processing techniques, such as tissue segmentation, require multiple MR contrasts as inputs, and each contrast is treated differently. Thus it is advantageous to automate the identification of image contrasts for various purposes, such as facilitating image processing pipelines, and managing and maintaining large databases via content-based image retrieval (CBIR). Most automated CBIR techniques focus on a two-step process: extracting features from data and classifying the image based on these features. We present a novel 3D deep convolutional neural network (CNN)-based method for MR image contrast classification. The proposed CNN automatically identifies the MR contrast of an input brain image volume. Specifically, we explored three classification problems: (1) identify T1-weighted (T1-w), T2-weighted (T2-w), and fluid-attenuated inversion recovery (FLAIR) contrasts, (2) identify pre vs post-contrast T1, (3) identify pre vs post-contrast FLAIR. A total of 3418 image volumes acquired from multiple sites and multiple scanners were used. To evaluate each task, the proposed model was trained on 2137 images and tested on the remaining 1281 images. Results showed that image volumes were correctly classified with 97.57% accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge