Colin B. Hansen
The Value of Nullspace Tuning Using Partial Label Information
Mar 17, 2020
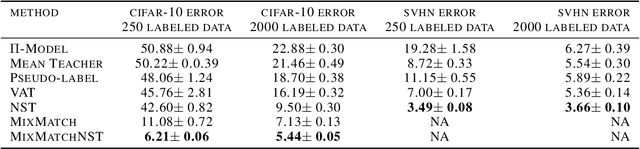


Abstract:In semi-supervised learning, information from unlabeled examples is used to improve the model learned from labeled examples. But in some learning problems, partial label information can be inferred from otherwise unlabeled examples and used to further improve the model. In particular, partial label information exists when subsets of training examples are known to have the same label, even though the label itself is missing. By encouraging a model to give the same label to all such examples, we can potentially improve its performance. We call this encouragement \emph{Nullspace Tuning} because the difference vector between any pair of examples with the same label should lie in the nullspace of a linear model. In this paper, we investigate the benefit of using partial label information using a careful comparison framework over well-characterized public datasets. We show that the additional information provided by partial labels reduces test error over good semi-supervised methods usually by a factor of 2, up to a factor of 5.5 in the best case. We also show that adding Nullspace Tuning to the newer and state-of-the-art MixMatch method decreases its test error by up to a factor of 1.8.
Deep Learning Captures More Accurate Diffusion Fiber Orientations Distributions than Constrained Spherical Deconvolution
Nov 13, 2019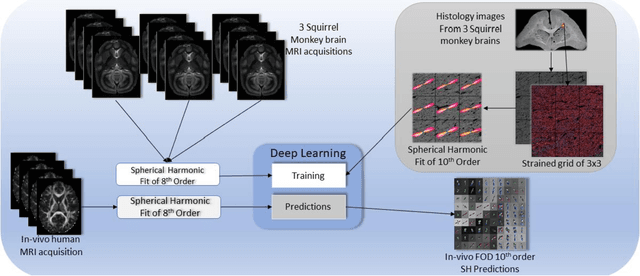
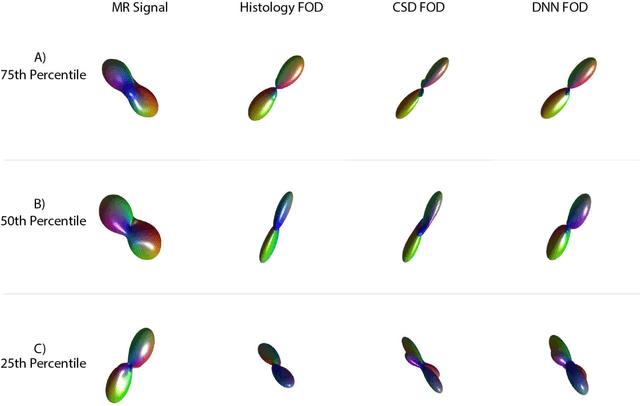
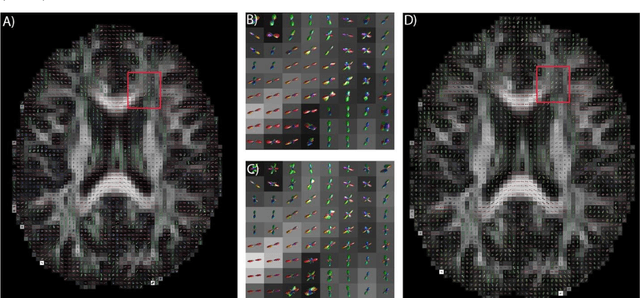
Abstract:Confocal histology provides an opportunity to establish intra-voxel fiber orientation distributions that can be used to quantitatively assess the biological relevance of diffusion weighted MRI models, e.g., constrained spherical deconvolution (CSD). Here, we apply deep learning to investigate the potential of single shell diffusion weighted MRI to explain histologically observed fiber orientation distributions (FOD) and compare the derived deep learning model with a leading CSD approach. This study (1) demonstrates that there exists additional information in the diffusion signal that is not currently exploited by CSD, and (2) provides an illustrative data-driven model that makes use of this information.
Enabling Multi-Shell b-Value Generalizability of Data-Driven Diffusion Models with Deep SHORE
Jul 15, 2019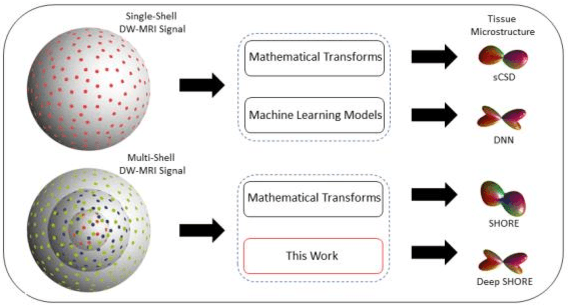
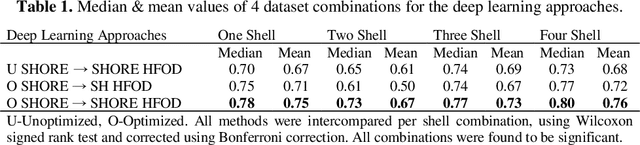
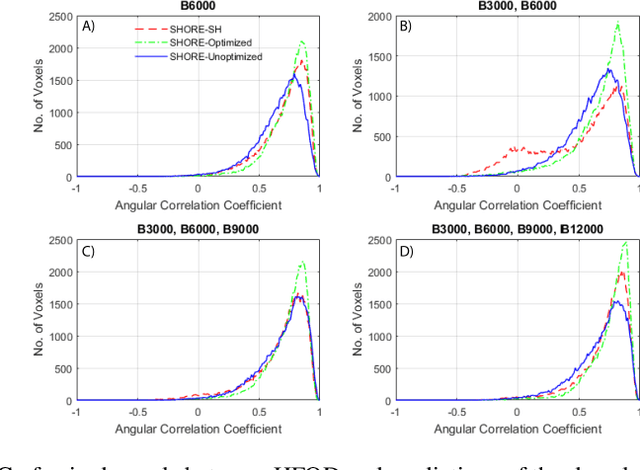
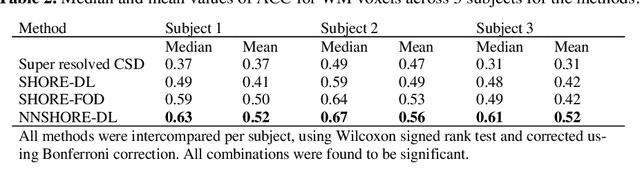
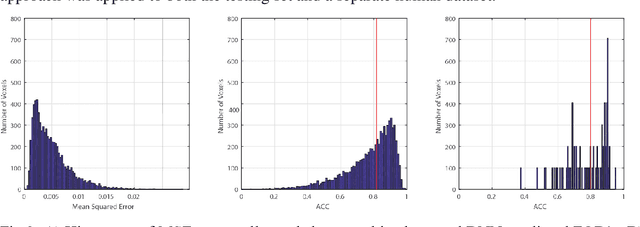
Abstract:Abstract. Intra-voxel models of the diffusion signal are essential for interpreting organization of the tissue environment at micrometer level with data at millimeter resolution. Recent advances in data driven methods have enabled direct compari-son and optimization of methods for in-vivo data with externally validated histological sections with both 2-D and 3-D histology. Yet, all existing methods make limiting assumptions of either (1) model-based linkages between b-values or (2) limited associations with single shell data. We generalize prior deep learning models that used single shell spherical harmonic transforms to integrate the re-cently developed simple harmonic oscillator reconstruction (SHORE) basis. To enable learning on the SHORE manifold, we present an alternative formulation of the fiber orientation distribution (FOD) object using the SHORE basis while rep-resenting the observed diffusion weighted data in the SHORE basis. To ensure consistency of hyper-parameter optimization for SHORE, we present our Deep SHORE approach to learn on a data-optimized manifold. Deep SHORE is evalu-ated with eight-fold cross-validation of a preclinical MRI-histology data with four b-values. Generalizability of in-vivo human data is evaluated on two separate 3T MRI scanners. Specificity in terms of angular correlation (ACC) with the preclinical data improved on single shell: 0.78 relative to 0.73 and 0.73, multi-shell: 0.80 relative to 0.74 (p < 0.001). In the in-vivo human data, Deep SHORE was more consistent across scanners with 0.63 relative to other multi-shell methods 0.39, 0.52 and 0.57 in terms of ACC. In conclusion, Deep SHORE is a promising method to enable data driven learning with DW-MRI under conditions with varying b-values, number of diffusion shells, and gradient directions per shell.
Inter-Scanner Harmonization of High Angular Resolution DW-MRI using Null Space Deep Learning
Oct 09, 2018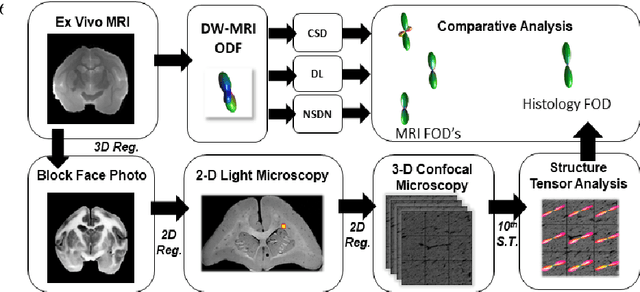
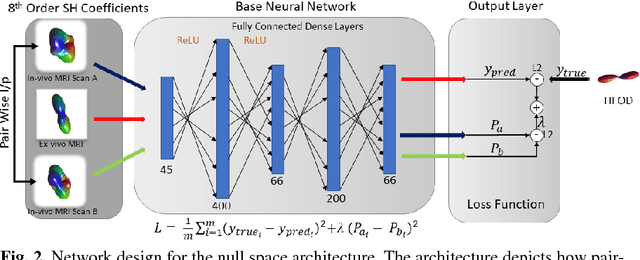

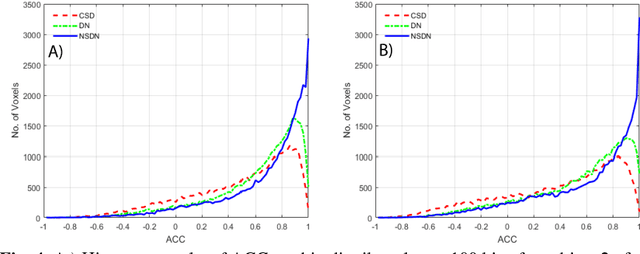
Abstract:Diffusion-weighted magnetic resonance imaging (DW-MRI) allows for non-invasive imaging of the local fiber architecture of the human brain at a millimetric scale. Multiple classical approaches have been proposed to detect both single (e.g., tensors) and multiple (e.g., constrained spherical deconvolution, CSD) fiber population orientations per voxel. However, existing techniques generally exhibit low reproducibility across MRI scanners. Herein, we propose a data-driven tech-nique using a neural network design which exploits two categories of data. First, training data were acquired on three squirrel monkey brains using ex-vivo DW-MRI and histology of the brain. Second, repeated scans of human subjects were acquired on two different scanners to augment the learning of the network pro-posed. To use these data, we propose a new network architecture, the null space deep network (NSDN), to simultaneously learn on traditional observed/truth pairs (e.g., MRI-histology voxels) along with repeated observations without a known truth (e.g., scan-rescan MRI). The NSDN was tested on twenty percent of the histology voxels that were kept completely blind to the network. NSDN significantly improved absolute performance relative to histology by 3.87% over CSD and 1.42% over a recently proposed deep neural network approach. More-over, it improved reproducibility on the paired data by 21.19% over CSD and 10.09% over a recently proposed deep approach. Finally, NSDN improved gen-eralizability of the model to a third in vivo human scanner (which was not used in training) by 16.08% over CSD and 10.41% over a recently proposed deep learn-ing approach. This work suggests that data-driven approaches for local fiber re-construction are more reproducible, informative and precise and offers a novel, practical method for determining these models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge