Thomas A. Lasko
Self-supervised learning of imaging and clinical signatures using a multimodal joint-embedding predictive architecture
Sep 18, 2025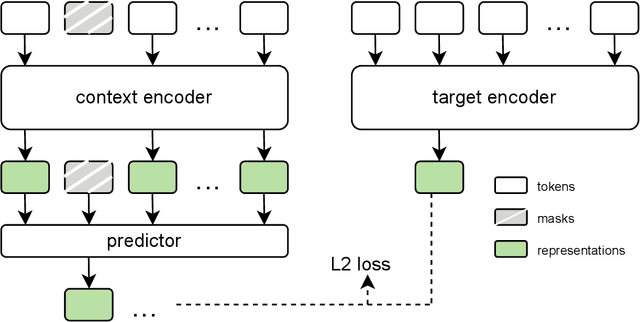

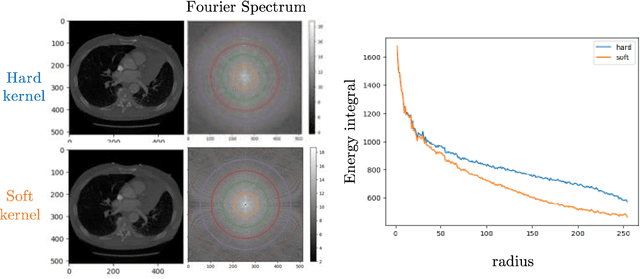
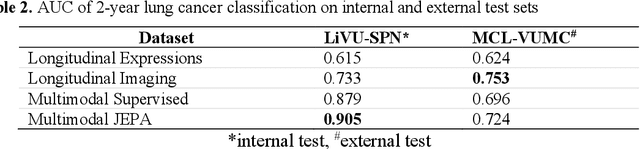
Abstract:The development of multimodal models for pulmonary nodule diagnosis is limited by the scarcity of labeled data and the tendency for these models to overfit on the training distribution. In this work, we leverage self-supervised learning from longitudinal and multimodal archives to address these challenges. We curate an unlabeled set of patients with CT scans and linked electronic health records from our home institution to power joint embedding predictive architecture (JEPA) pretraining. After supervised finetuning, we show that our approach outperforms an unregularized multimodal model and imaging-only model in an internal cohort (ours: 0.91, multimodal: 0.88, imaging-only: 0.73 AUC), but underperforms in an external cohort (ours: 0.72, imaging-only: 0.75 AUC). We develop a synthetic environment that characterizes the context in which JEPA may underperform. This work innovates an approach that leverages unlabeled multimodal medical archives to improve predictive models and demonstrates its advantages and limitations in pulmonary nodule diagnosis.
Lifespan Pancreas Morphology for Control vs Type 2 Diabetes using AI on Largescale Clinical Imaging
Aug 20, 2025Abstract:Purpose: Understanding how the pancreas changes is critical for detecting deviations in type 2 diabetes and other pancreatic disease. We measure pancreas size and shape using morphological measurements from ages 0 to 90. Our goals are to 1) identify reliable clinical imaging modalities for AI-based pancreas measurement, 2) establish normative morphological aging trends, and 3) detect potential deviations in type 2 diabetes. Approach: We analyzed a clinically acquired dataset of 2533 patients imaged with abdominal CT or MRI. We resampled the scans to 3mm isotropic resolution, segmented the pancreas using automated methods, and extracted 13 morphological pancreas features across the lifespan. First, we assessed CT and MRI measurements to determine which modalities provide consistent lifespan trends. Second, we characterized distributions of normative morphological patterns stratified by age group and sex. Third, we used GAMLSS regression to model pancreas morphology trends in 1350 patients matched for age, sex, and type 2 diabetes status to identify any deviations from normative aging associated with type 2 diabetes. Results: When adjusting for confounders, the aging trends for 10 of 13 morphological features were significantly different between patients with type 2 diabetes and non-diabetic controls (p < 0.05 after multiple comparisons corrections). Additionally, MRI appeared to yield different pancreas measurements than CT using our AI-based method. Conclusions: We provide lifespan trends demonstrating that the size and shape of the pancreas is altered in type 2 diabetes using 675 control patients and 675 diabetes patients. Moreover, our findings reinforce that the pancreas is smaller in type 2 diabetes. Additionally, we contribute a reference of lifespan pancreas morphology from a large cohort of non-diabetic control patients in a clinical setting.
Cryptogenic stroke and migraine: using probabilistic independence and machine learning to uncover latent sources of disease from the electronic health record
Apr 22, 2025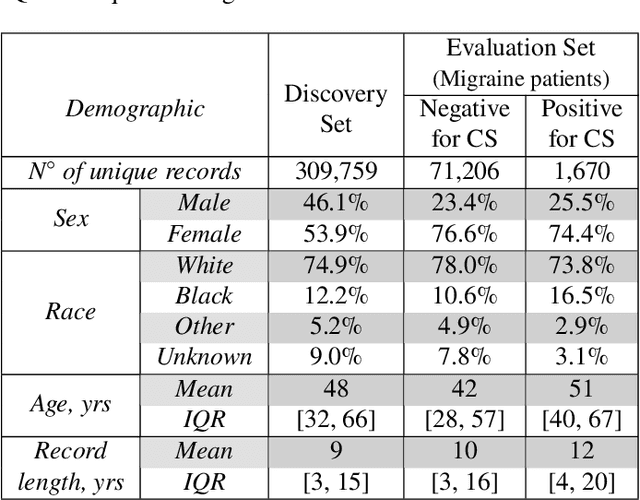
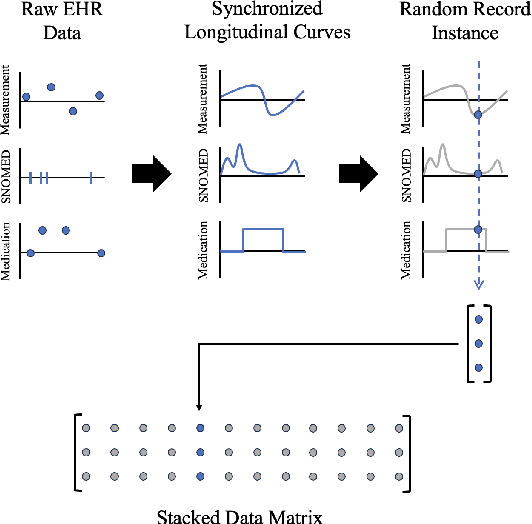
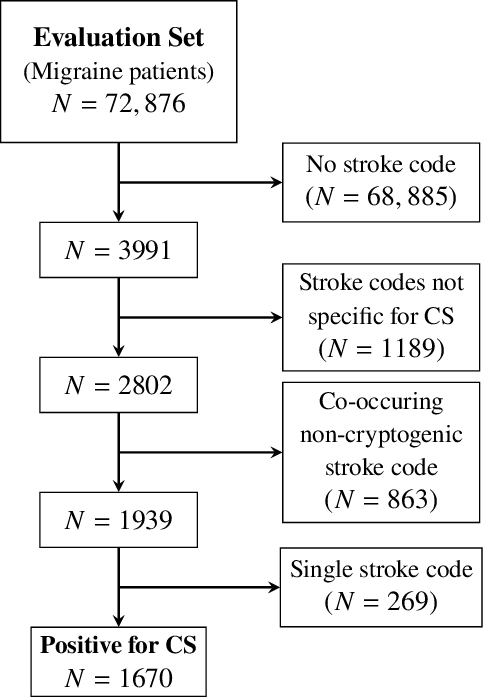
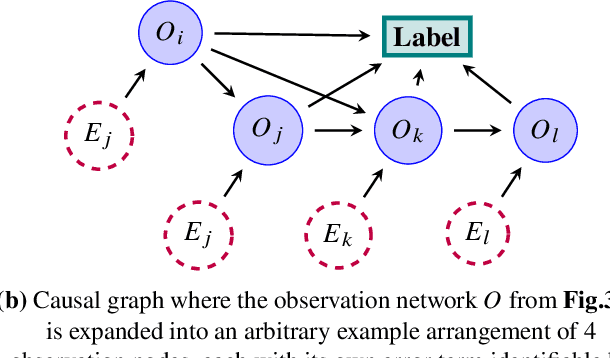
Abstract:Migraine is a common but complex neurological disorder that doubles the lifetime risk of cryptogenic stroke (CS). However, this relationship remains poorly characterized, and few clinical guidelines exist to reduce this associated risk. We therefore propose a data-driven approach to extract probabilistically-independent sources from electronic health record (EHR) data and create a 10-year risk-predictive model for CS in migraine patients. These sources represent external latent variables acting on the causal graph constructed from the EHR data and approximate root causes of CS in our population. A random forest model trained on patient expressions of these sources demonstrated good accuracy (ROC 0.771) and identified the top 10 most predictive sources of CS in migraine patients. These sources revealed that pharmacologic interventions were the most important factor in minimizing CS risk in our population and identified a factor related to allergic rhinitis as a potential causative source of CS in migraine patients.
A data-driven approach to discover and quantify systemic lupus erythematosus etiological heterogeneity from electronic health records
Jan 13, 2025Abstract:Systemic lupus erythematosus (SLE) is a complex heterogeneous disease with many manifestational facets. We propose a data-driven approach to discover probabilistic independent sources from multimodal imperfect EHR data. These sources represent exogenous variables in the data generation process causal graph that estimate latent root causes of the presence of SLE in the health record. We objectively evaluated the sources against the original variables from which they were discovered by training supervised models to discriminate SLE from negative health records using a reduced set of labelled instances. We found 19 predictive sources with high clinical validity and whose EHR signatures define independent factors of SLE heterogeneity. Using the sources as input patient data representation enables models to provide with rich explanations that better capture the clinical reasons why a particular record is (not) an SLE case. Providers may be willing to trade patient-level interpretability for discrimination especially in challenging cases.
Unsupervised Discovery of Clinical Disease Signatures Using Probabilistic Independence
Feb 08, 2024Abstract:Insufficiently precise diagnosis of clinical disease is likely responsible for many treatment failures, even for common conditions and treatments. With a large enough dataset, it may be possible to use unsupervised machine learning to define clinical disease patterns more precisely. We present an approach to learning these patterns by using probabilistic independence to disentangle the imprint on the medical record of causal latent sources of disease. We inferred a broad set of 2000 clinical signatures of latent sources from 9195 variables in 269,099 Electronic Health Records. The learned signatures produced better discrimination than the original variables in a lung cancer prediction task unknown to the inference algorithm, predicting 3-year malignancy in patients with no history of cancer before a solitary lung nodule was discovered. More importantly, the signatures' greater explanatory power identified pre-nodule signatures of apparently undiagnosed cancer in many of those patients.
Why Do Clinical Probabilistic Models Fail To Transport Between Sites?
Nov 08, 2023Abstract:The rising popularity of artificial intelligence in healthcare is highlighting the problem that a computational model achieving super-human clinical performance at its training sites may perform substantially worse at new sites. In this perspective, we present common sources for this failure to transport, which we divide into sources under the control of the experimenter and sources inherent to the clinical data-generating process. Of the inherent sources we look a little deeper into site-specific clinical practices that can affect the data distribution, and propose a potential solution intended to isolate the imprint of those practices on the data from the patterns of disease cause and effect that are the usual target of clinical models.
Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures From Routine EHRs for Pulmonary Nodule Classification
Apr 10, 2023



Abstract:The accuracy of predictive models for solitary pulmonary nodule (SPN) diagnosis can be greatly increased by incorporating repeat imaging and medical context, such as electronic health records (EHRs). However, clinically routine modalities such as imaging and diagnostic codes can be asynchronous and irregularly sampled over different time scales which are obstacles to longitudinal multimodal learning. In this work, we propose a transformer-based multimodal strategy to integrate repeat imaging with longitudinal clinical signatures from routinely collected EHRs for SPN classification. We perform unsupervised disentanglement of latent clinical signatures and leverage time-distance scaled self-attention to jointly learn from clinical signatures expressions and chest computed tomography (CT) scans. Our classifier is pretrained on 2,668 scans from a public dataset and 1,149 subjects with longitudinal chest CTs, billing codes, medications, and laboratory tests from EHRs of our home institution. Evaluation on 227 subjects with challenging SPNs revealed a significant AUC improvement over a longitudinal multimodal baseline (0.824 vs 0.752 AUC), as well as improvements over a single cross-section multimodal scenario (0.809 AUC) and a longitudinal imaging-only scenario (0.741 AUC). This work demonstrates significant advantages with a novel approach for co-learning longitudinal imaging and non-imaging phenotypes with transformers.
Sample-Specific Root Causal Inference with Latent Variables
Oct 27, 2022Abstract:Root causal analysis seeks to identify the set of initial perturbations that induce an unwanted outcome. In prior work, we defined sample-specific root causes of disease using exogenous error terms that predict a diagnosis in a structural equation model. We rigorously quantified predictivity using Shapley values. However, the associated algorithms for inferring root causes assume no latent confounding. We relax this assumption by permitting confounding among the predictors. We then introduce a corresponding procedure called Extract Errors with Latents (EEL) for recovering the error terms up to contamination by vertices on certain paths under the linear non-Gaussian acyclic model. EEL also identifies the smallest sets of dependent errors for fast computation of the Shapley values. The algorithm bypasses the hard problem of estimating the underlying causal graph in both cases. Experiments highlight the superior accuracy and robustness of EEL relative to its predecessors.
UNesT: Local Spatial Representation Learning with Hierarchical Transformer for Efficient Medical Segmentation
Sep 28, 2022


Abstract:Transformer-based models, capable of learning better global dependencies, have recently demonstrated exceptional representation learning capabilities in computer vision and medical image analysis. Transformer reformats the image into separate patches and realize global communication via the self-attention mechanism. However, positional information between patches is hard to preserve in such 1D sequences, and loss of it can lead to sub-optimal performance when dealing with large amounts of heterogeneous tissues of various sizes in 3D medical image segmentation. Additionally, current methods are not robust and efficient for heavy-duty medical segmentation tasks such as predicting a large number of tissue classes or modeling globally inter-connected tissues structures. Inspired by the nested hierarchical structures in vision transformer, we proposed a novel 3D medical image segmentation method (UNesT), employing a simplified and faster-converging transformer encoder design that achieves local communication among spatially adjacent patch sequences by aggregating them hierarchically. We extensively validate our method on multiple challenging datasets, consisting anatomies of 133 structures in brain, 14 organs in abdomen, 4 hierarchical components in kidney, and inter-connected kidney tumors). We show that UNesT consistently achieves state-of-the-art performance and evaluate its generalizability and data efficiency. Particularly, the model achieves whole brain segmentation task complete ROI with 133 tissue classes in single network, outperforms prior state-of-the-art method SLANT27 ensembled with 27 network tiles, our model performance increases the mean DSC score of the publicly available Colin and CANDI dataset from 0.7264 to 0.7444 and from 0.6968 to 0.7025, respectively.
Time-distance vision transformers in lung cancer diagnosis from longitudinal computed tomography
Sep 04, 2022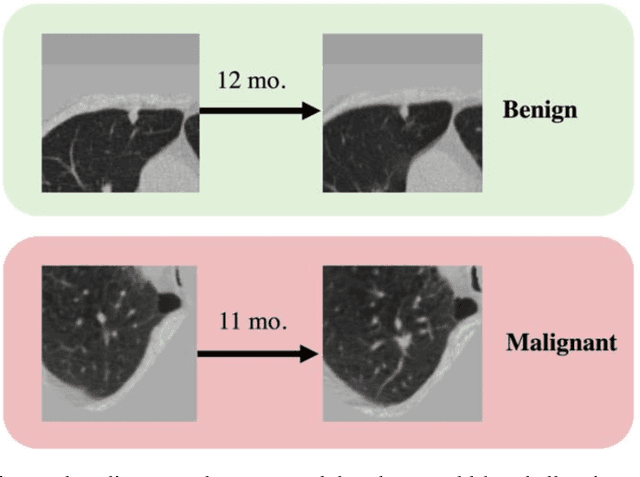

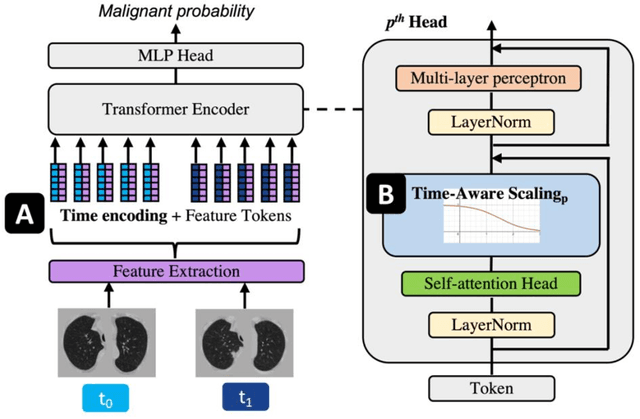

Abstract:Features learned from single radiologic images are unable to provide information about whether and how much a lesion may be changing over time. Time-dependent features computed from repeated images can capture those changes and help identify malignant lesions by their temporal behavior. However, longitudinal medical imaging presents the unique challenge of sparse, irregular time intervals in data acquisition. While self-attention has been shown to be a versatile and efficient learning mechanism for time series and natural images, its potential for interpreting temporal distance between sparse, irregularly sampled spatial features has not been explored. In this work, we propose two interpretations of a time-distance vision transformer (ViT) by using (1) vector embeddings of continuous time and (2) a temporal emphasis model to scale self-attention weights. The two algorithms are evaluated based on benign versus malignant lung cancer discrimination of synthetic pulmonary nodules and lung screening computed tomography studies from the National Lung Screening Trial (NLST). Experiments evaluating the time-distance ViTs on synthetic nodules show a fundamental improvement in classifying irregularly sampled longitudinal images when compared to standard ViTs. In cross-validation on screening chest CTs from the NLST, our methods (0.785 and 0.786 AUC respectively) significantly outperform a cross-sectional approach (0.734 AUC) and match the discriminative performance of the leading longitudinal medical imaging algorithm (0.779 AUC) on benign versus malignant classification. This work represents the first self-attention-based framework for classifying longitudinal medical images. Our code is available at https://github.com/tom1193/time-distance-transformer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge