Kim Sandler
Time-distance vision transformers in lung cancer diagnosis from longitudinal computed tomography
Sep 04, 2022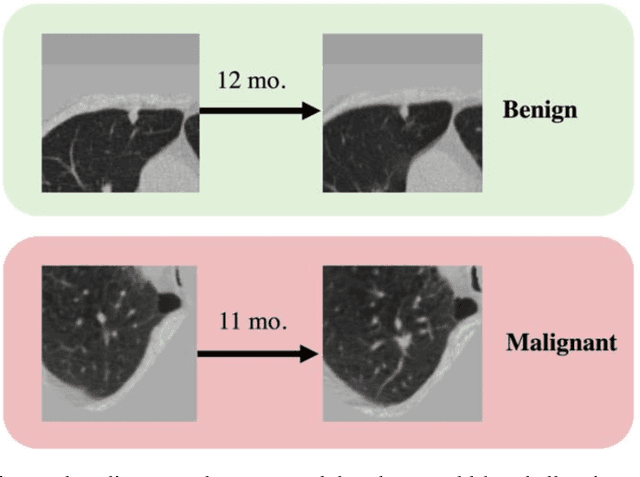

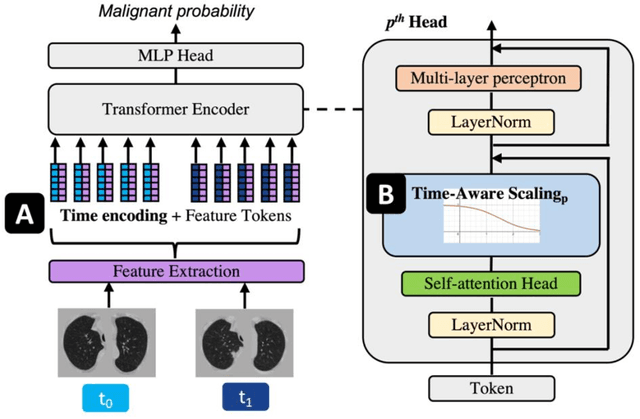

Abstract:Features learned from single radiologic images are unable to provide information about whether and how much a lesion may be changing over time. Time-dependent features computed from repeated images can capture those changes and help identify malignant lesions by their temporal behavior. However, longitudinal medical imaging presents the unique challenge of sparse, irregular time intervals in data acquisition. While self-attention has been shown to be a versatile and efficient learning mechanism for time series and natural images, its potential for interpreting temporal distance between sparse, irregularly sampled spatial features has not been explored. In this work, we propose two interpretations of a time-distance vision transformer (ViT) by using (1) vector embeddings of continuous time and (2) a temporal emphasis model to scale self-attention weights. The two algorithms are evaluated based on benign versus malignant lung cancer discrimination of synthetic pulmonary nodules and lung screening computed tomography studies from the National Lung Screening Trial (NLST). Experiments evaluating the time-distance ViTs on synthetic nodules show a fundamental improvement in classifying irregularly sampled longitudinal images when compared to standard ViTs. In cross-validation on screening chest CTs from the NLST, our methods (0.785 and 0.786 AUC respectively) significantly outperform a cross-sectional approach (0.734 AUC) and match the discriminative performance of the leading longitudinal medical imaging algorithm (0.779 AUC) on benign versus malignant classification. This work represents the first self-attention-based framework for classifying longitudinal medical images. Our code is available at https://github.com/tom1193/time-distance-transformer.
A Comparative Study of Confidence Calibration in Deep Learning: From Computer Vision to Medical Imaging
Jun 17, 2022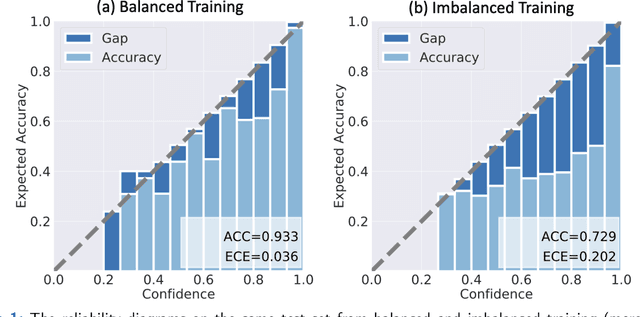
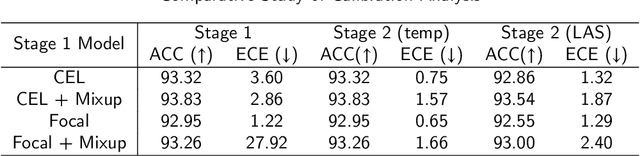
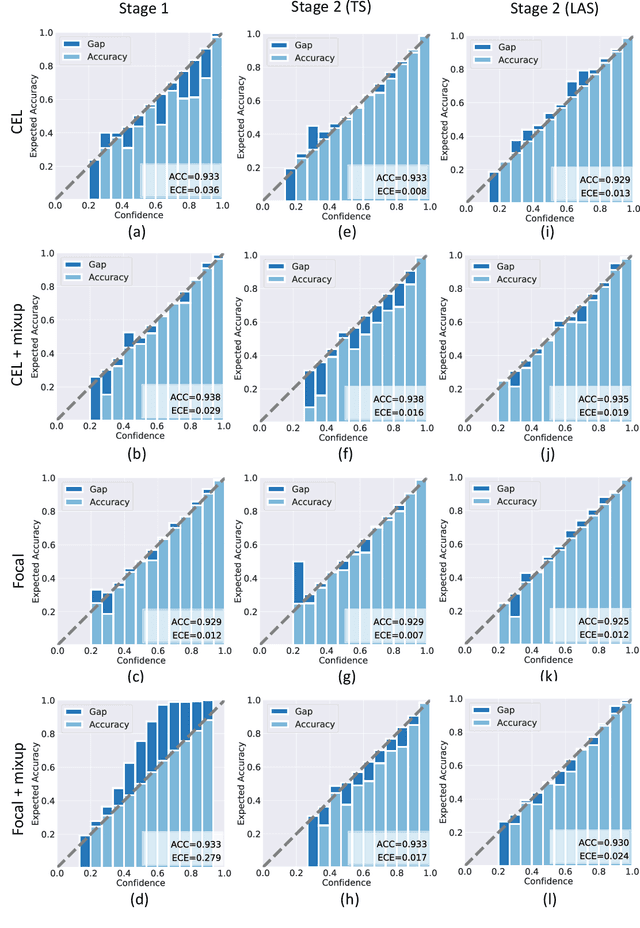
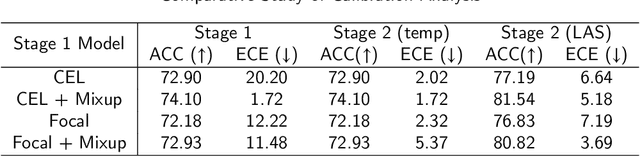
Abstract:Although deep learning prediction models have been successful in the discrimination of different classes, they can often suffer from poor calibration across challenging domains including healthcare. Moreover, the long-tail distribution poses great challenges in deep learning classification problems including clinical disease prediction. There are approaches proposed recently to calibrate deep prediction in computer vision, but there are no studies found to demonstrate how the representative models work in different challenging contexts. In this paper, we bridge the confidence calibration from computer vision to medical imaging with a comparative study of four high-impact calibration models. Our studies are conducted in different contexts (natural image classification and lung cancer risk estimation) including in balanced vs. imbalanced training sets and in computer vision vs. medical imaging. Our results support key findings: (1) We achieve new conclusions which are not studied under different learning contexts, e.g., combining two calibration models that both mitigate the overconfident prediction can lead to under-confident prediction, and simpler calibration models from the computer vision domain tend to be more generalizable to medical imaging. (2) We highlight the gap between general computer vision tasks and medical imaging prediction, e.g., calibration methods ideal for general computer vision tasks may in fact damage the calibration of medical imaging prediction. (3) We also reinforce previous conclusions in natural image classification settings. We believe that this study has merits to guide readers to choose calibration models and understand gaps between general computer vision and medical imaging domains.
Lung Cancer Risk Estimation with Incomplete Data: A Joint Missing Imputation Perspective
Jul 25, 2021



Abstract:Data from multi-modality provide complementary information in clinical prediction, but missing data in clinical cohorts limits the number of subjects in multi-modal learning context. Multi-modal missing imputation is challenging with existing methods when 1) the missing data span across heterogeneous modalities (e.g., image vs. non-image); or 2) one modality is largely missing. In this paper, we address imputation of missing data by modeling the joint distribution of multi-modal data. Motivated by partial bidirectional generative adversarial net (PBiGAN), we propose a new Conditional PBiGAN (C-PBiGAN) method that imputes one modality combining the conditional knowledge from another modality. Specifically, C-PBiGAN introduces a conditional latent space in a missing imputation framework that jointly encodes the available multi-modal data, along with a class regularization loss on imputed data to recover discriminative information. To our knowledge, it is the first generative adversarial model that addresses multi-modal missing imputation by modeling the joint distribution of image and non-image data. We validate our model with both the national lung screening trial (NLST) dataset and an external clinical validation cohort. The proposed C-PBiGAN achieves significant improvements in lung cancer risk estimation compared with representative imputation methods (e.g., AUC values increase in both NLST (+2.9\%) and in-house dataset (+4.3\%) compared with PBiGAN, p$<$0.05).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge