Kaiwen Xu
Multipath cycleGAN for harmonization of paired and unpaired low-dose lung computed tomography reconstruction kernels
May 28, 2025Abstract:Reconstruction kernels in computed tomography (CT) affect spatial resolution and noise characteristics, introducing systematic variability in quantitative imaging measurements such as emphysema quantification. Choosing an appropriate kernel is therefore essential for consistent quantitative analysis. We propose a multipath cycleGAN model for CT kernel harmonization, trained on a mixture of paired and unpaired data from a low-dose lung cancer screening cohort. The model features domain-specific encoders and decoders with a shared latent space and uses discriminators tailored for each domain.We train the model on 42 kernel combinations using 100 scans each from seven representative kernels in the National Lung Screening Trial (NLST) dataset. To evaluate performance, 240 scans from each kernel are harmonized to a reference soft kernel, and emphysema is quantified before and after harmonization. A general linear model assesses the impact of age, sex, smoking status, and kernel on emphysema. We also evaluate harmonization from soft kernels to a reference hard kernel. To assess anatomical consistency, we compare segmentations of lung vessels, muscle, and subcutaneous adipose tissue generated by TotalSegmentator between harmonized and original images. Our model is benchmarked against traditional and switchable cycleGANs. For paired kernels, our approach reduces bias in emphysema scores, as seen in Bland-Altman plots (p<0.05). For unpaired kernels, harmonization eliminates confounding differences in emphysema (p>0.05). High Dice scores confirm preservation of muscle and fat anatomy, while lung vessel overlap remains reasonable. Overall, our shared latent space multipath cycleGAN enables robust harmonization across paired and unpaired CT kernels, improving emphysema quantification and preserving anatomical fidelity.
Investigating the impact of kernel harmonization and deformable registration on inspiratory and expiratory chest CT images for people with COPD
Feb 07, 2025



Abstract:Paired inspiratory-expiratory CT scans enable the quantification of gas trapping due to small airway disease and emphysema by analyzing lung tissue motion in COPD patients. Deformable image registration of these scans assesses regional lung volumetric changes. However, variations in reconstruction kernels between paired scans introduce errors in quantitative analysis. This work proposes a two-stage pipeline to harmonize reconstruction kernels and perform deformable image registration using data acquired from the COPDGene study. We use a cycle generative adversarial network (GAN) to harmonize inspiratory scans reconstructed with a hard kernel (BONE) to match expiratory scans reconstructed with a soft kernel (STANDARD). We then deformably register the expiratory scans to inspiratory scans. We validate harmonization by measuring emphysema using a publicly available segmentation algorithm before and after harmonization. Results show harmonization significantly reduces emphysema measurement inconsistencies, decreasing median emphysema scores from 10.479% to 3.039%, with a reference median score of 1.305% from the STANDARD kernel as the target. Registration accuracy is evaluated via Dice overlap between emphysema regions on inspiratory, expiratory, and deformed images. The Dice coefficient between inspiratory emphysema masks and deformably registered emphysema masks increases significantly across registration stages (p<0.001). Additionally, we demonstrate that deformable registration is robust to kernel variations.
Pitfalls of defacing whole-head MRI: re-identification risk with diffusion models and compromised research potential
Jan 31, 2025Abstract:Defacing is often applied to head magnetic resonance image (MRI) datasets prior to public release to address privacy concerns. The alteration of facial and nearby voxels has provoked discussions about the true capability of these techniques to ensure privacy as well as their impact on downstream tasks. With advancements in deep generative models, the extent to which defacing can protect privacy is uncertain. Additionally, while the altered voxels are known to contain valuable anatomical information, their potential to support research beyond the anatomical regions directly affected by defacing remains uncertain. To evaluate these considerations, we develop a refacing pipeline that recovers faces in defaced head MRIs using cascaded diffusion probabilistic models (DPMs). The DPMs are trained on images from 180 subjects and tested on images from 484 unseen subjects, 469 of whom are from a different dataset. To assess whether the altered voxels in defacing contain universally useful information, we also predict computed tomography (CT)-derived skeletal muscle radiodensity from facial voxels in both defaced and original MRIs. The results show that DPMs can generate high-fidelity faces that resemble the original faces from defaced images, with surface distances to the original faces significantly smaller than those of a population average face (p < 0.05). This performance also generalizes well to previously unseen datasets. For skeletal muscle radiodensity predictions, using defaced images results in significantly weaker Spearman's rank correlation coefficients compared to using original images (p < 10-4). For shin muscle, the correlation is statistically significant (p < 0.05) when using original images but not statistically significant (p > 0.05) when any defacing method is applied, suggesting that defacing might not only fail to protect privacy but also eliminate valuable information.
Synthesizing Proton-Density Fat Fraction and $R_2^*$ from 2-point Dixon MRI with Generative Machine Learning
Oct 15, 2024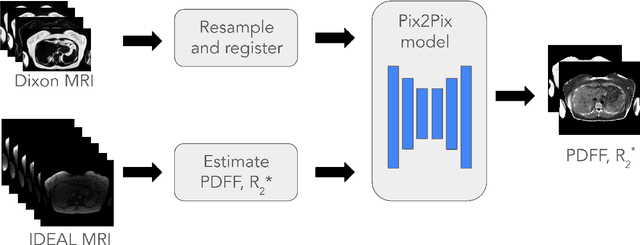



Abstract:Magnetic Resonance Imaging (MRI) is the gold standard for measuring fat and iron content non-invasively in the body via measures known as Proton Density Fat Fraction (PDFF) and $R_2^*$, respectively. However, conventional PDFF and $R_2^*$ quantification methods operate on MR images voxel-wise and require at least three measurements to estimate three quantities: water, fat, and $R_2^*$. Alternatively, the two-point Dixon MRI protocol is widely used and fast because it acquires only two measurements; however, these cannot be used to estimate three quantities voxel-wise. Leveraging the fact that neighboring voxels have similar values, we propose using a generative machine learning approach to learn PDFF and $R_2^*$ from Dixon MRI. We use paired Dixon-IDEAL data from UK Biobank in the liver and a Pix2Pix conditional GAN to demonstrate the first large-scale $R_2^*$ imputation from two-point Dixon MRIs. Using our proposed approach, we synthesize PDFF and $R_2^*$ maps that show significantly greater correlation with ground-truth than conventional voxel-wise baselines.
Inter-vendor harmonization of Computed Tomography (CT) reconstruction kernels using unpaired image translation
Sep 22, 2023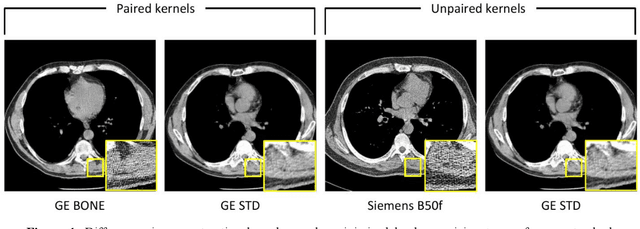
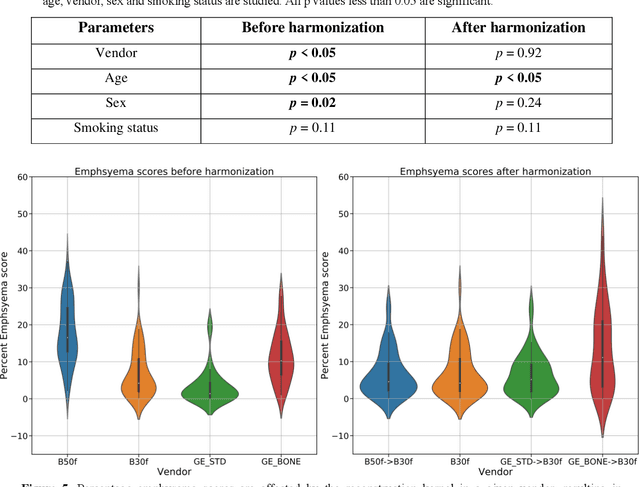
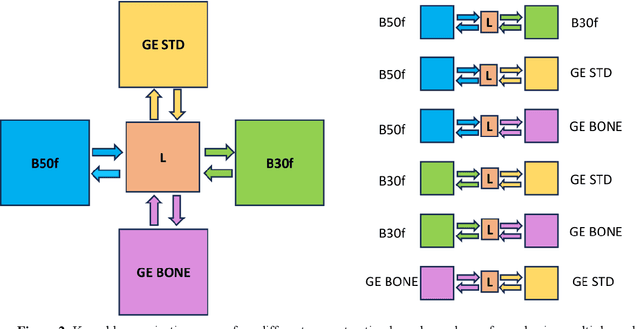
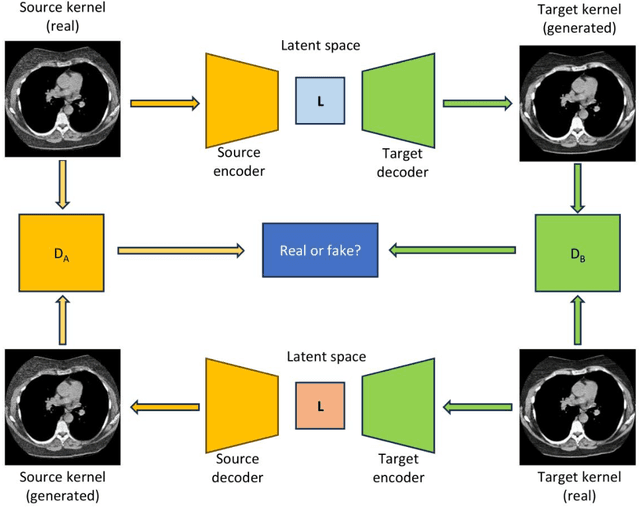
Abstract:The reconstruction kernel in computed tomography (CT) generation determines the texture of the image. Consistency in reconstruction kernels is important as the underlying CT texture can impact measurements during quantitative image analysis. Harmonization (i.e., kernel conversion) minimizes differences in measurements due to inconsistent reconstruction kernels. Existing methods investigate harmonization of CT scans in single or multiple manufacturers. However, these methods require paired scans of hard and soft reconstruction kernels that are spatially and anatomically aligned. Additionally, a large number of models need to be trained across different kernel pairs within manufacturers. In this study, we adopt an unpaired image translation approach to investigate harmonization between and across reconstruction kernels from different manufacturers by constructing a multipath cycle generative adversarial network (GAN). We use hard and soft reconstruction kernels from the Siemens and GE vendors from the National Lung Screening Trial dataset. We use 50 scans from each reconstruction kernel and train a multipath cycle GAN. To evaluate the effect of harmonization on the reconstruction kernels, we harmonize 50 scans each from Siemens hard kernel, GE soft kernel and GE hard kernel to a reference Siemens soft kernel (B30f) and evaluate percent emphysema. We fit a linear model by considering the age, smoking status, sex and vendor and perform an analysis of variance (ANOVA) on the emphysema scores. Our approach minimizes differences in emphysema measurement and highlights the impact of age, sex, smoking status and vendor on emphysema quantification.
Statistically Significant Concept-based Explanation of Image Classifiers via Model Knockoffs
May 31, 2023



Abstract:A concept-based classifier can explain the decision process of a deep learning model by human-understandable concepts in image classification problems. However, sometimes concept-based explanations may cause false positives, which misregards unrelated concepts as important for the prediction task. Our goal is to find the statistically significant concept for classification to prevent misinterpretation. In this study, we propose a method using a deep learning model to learn the image concept and then using the Knockoff samples to select the important concepts for prediction by controlling the False Discovery Rate (FDR) under a certain value. We evaluate the proposed method in our synthetic and real data experiments. Also, it shows that our method can control the FDR properly while selecting highly interpretable concepts to improve the trustworthiness of the model.
Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures From Routine EHRs for Pulmonary Nodule Classification
Apr 10, 2023



Abstract:The accuracy of predictive models for solitary pulmonary nodule (SPN) diagnosis can be greatly increased by incorporating repeat imaging and medical context, such as electronic health records (EHRs). However, clinically routine modalities such as imaging and diagnostic codes can be asynchronous and irregularly sampled over different time scales which are obstacles to longitudinal multimodal learning. In this work, we propose a transformer-based multimodal strategy to integrate repeat imaging with longitudinal clinical signatures from routinely collected EHRs for SPN classification. We perform unsupervised disentanglement of latent clinical signatures and leverage time-distance scaled self-attention to jointly learn from clinical signatures expressions and chest computed tomography (CT) scans. Our classifier is pretrained on 2,668 scans from a public dataset and 1,149 subjects with longitudinal chest CTs, billing codes, medications, and laboratory tests from EHRs of our home institution. Evaluation on 227 subjects with challenging SPNs revealed a significant AUC improvement over a longitudinal multimodal baseline (0.824 vs 0.752 AUC), as well as improvements over a single cross-section multimodal scenario (0.809 AUC) and a longitudinal imaging-only scenario (0.741 AUC). This work demonstrates significant advantages with a novel approach for co-learning longitudinal imaging and non-imaging phenotypes with transformers.
Zero-shot CT Field-of-view Completion with Unconditional Generative Diffusion Prior
Apr 07, 2023Abstract:Anatomically consistent field-of-view (FOV) completion to recover truncated body sections has important applications in quantitative analyses of computed tomography (CT) with limited FOV. Existing solution based on conditional generative models relies on the fidelity of synthetic truncation patterns at training phase, which poses limitations for the generalizability of the method to potential unknown types of truncation. In this study, we evaluate a zero-shot method based on a pretrained unconditional generative diffusion prior, where truncation pattern with arbitrary forms can be specified at inference phase. In evaluation on simulated chest CT slices with synthetic FOV truncation, the method is capable of recovering anatomically consistent body sections and subcutaneous adipose tissue measurement error caused by FOV truncation. However, the correction accuracy is inferior to the conditionally trained counterpart.
Single Slice Thigh CT Muscle Group Segmentation with Domain Adaptation and Self-Training
Nov 30, 2022Abstract:Objective: Thigh muscle group segmentation is important for assessment of muscle anatomy, metabolic disease and aging. Many efforts have been put into quantifying muscle tissues with magnetic resonance (MR) imaging including manual annotation of individual muscles. However, leveraging publicly available annotations in MR images to achieve muscle group segmentation on single slice computed tomography (CT) thigh images is challenging. Method: We propose an unsupervised domain adaptation pipeline with self-training to transfer labels from 3D MR to single CT slice. First, we transform the image appearance from MR to CT with CycleGAN and feed the synthesized CT images to a segmenter simultaneously. Single CT slices are divided into hard and easy cohorts based on the entropy of pseudo labels inferenced by the segmenter. After refining easy cohort pseudo labels based on anatomical assumption, self-training with easy and hard splits is applied to fine tune the segmenter. Results: On 152 withheld single CT thigh images, the proposed pipeline achieved a mean Dice of 0.888(0.041) across all muscle groups including sartorius, hamstrings, quadriceps femoris and gracilis. muscles Conclusion: To our best knowledge, this is the first pipeline to achieve thigh imaging domain adaptation from MR to CT. The proposed pipeline is effective and robust in extracting muscle groups on 2D single slice CT thigh images.The container is available for public use at https://github.com/MASILab/DA_CT_muscle_seg
Time-distance vision transformers in lung cancer diagnosis from longitudinal computed tomography
Sep 04, 2022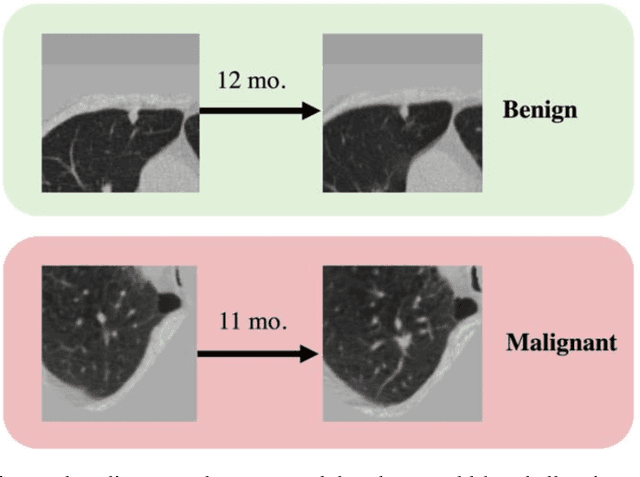

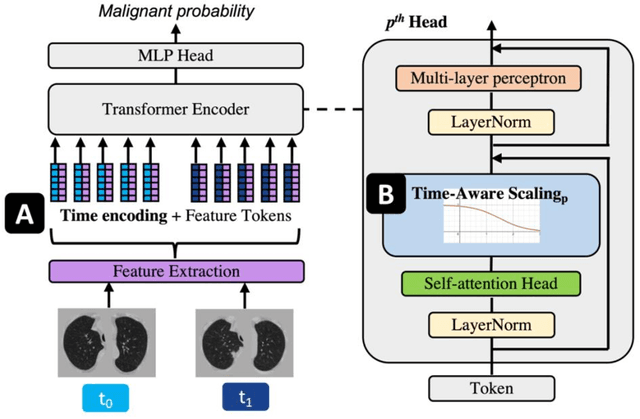

Abstract:Features learned from single radiologic images are unable to provide information about whether and how much a lesion may be changing over time. Time-dependent features computed from repeated images can capture those changes and help identify malignant lesions by their temporal behavior. However, longitudinal medical imaging presents the unique challenge of sparse, irregular time intervals in data acquisition. While self-attention has been shown to be a versatile and efficient learning mechanism for time series and natural images, its potential for interpreting temporal distance between sparse, irregularly sampled spatial features has not been explored. In this work, we propose two interpretations of a time-distance vision transformer (ViT) by using (1) vector embeddings of continuous time and (2) a temporal emphasis model to scale self-attention weights. The two algorithms are evaluated based on benign versus malignant lung cancer discrimination of synthetic pulmonary nodules and lung screening computed tomography studies from the National Lung Screening Trial (NLST). Experiments evaluating the time-distance ViTs on synthetic nodules show a fundamental improvement in classifying irregularly sampled longitudinal images when compared to standard ViTs. In cross-validation on screening chest CTs from the NLST, our methods (0.785 and 0.786 AUC respectively) significantly outperform a cross-sectional approach (0.734 AUC) and match the discriminative performance of the leading longitudinal medical imaging algorithm (0.779 AUC) on benign versus malignant classification. This work represents the first self-attention-based framework for classifying longitudinal medical images. Our code is available at https://github.com/tom1193/time-distance-transformer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge