Michael Kammer
Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures From Routine EHRs for Pulmonary Nodule Classification
Apr 10, 2023



Abstract:The accuracy of predictive models for solitary pulmonary nodule (SPN) diagnosis can be greatly increased by incorporating repeat imaging and medical context, such as electronic health records (EHRs). However, clinically routine modalities such as imaging and diagnostic codes can be asynchronous and irregularly sampled over different time scales which are obstacles to longitudinal multimodal learning. In this work, we propose a transformer-based multimodal strategy to integrate repeat imaging with longitudinal clinical signatures from routinely collected EHRs for SPN classification. We perform unsupervised disentanglement of latent clinical signatures and leverage time-distance scaled self-attention to jointly learn from clinical signatures expressions and chest computed tomography (CT) scans. Our classifier is pretrained on 2,668 scans from a public dataset and 1,149 subjects with longitudinal chest CTs, billing codes, medications, and laboratory tests from EHRs of our home institution. Evaluation on 227 subjects with challenging SPNs revealed a significant AUC improvement over a longitudinal multimodal baseline (0.824 vs 0.752 AUC), as well as improvements over a single cross-section multimodal scenario (0.809 AUC) and a longitudinal imaging-only scenario (0.741 AUC). This work demonstrates significant advantages with a novel approach for co-learning longitudinal imaging and non-imaging phenotypes with transformers.
A Comparative Study of Confidence Calibration in Deep Learning: From Computer Vision to Medical Imaging
Jun 17, 2022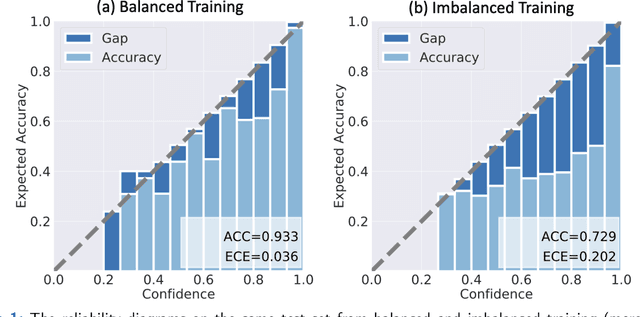
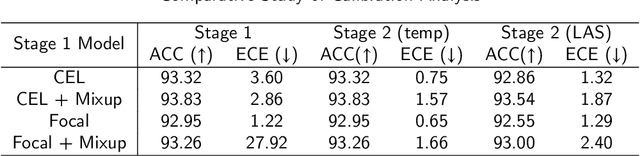
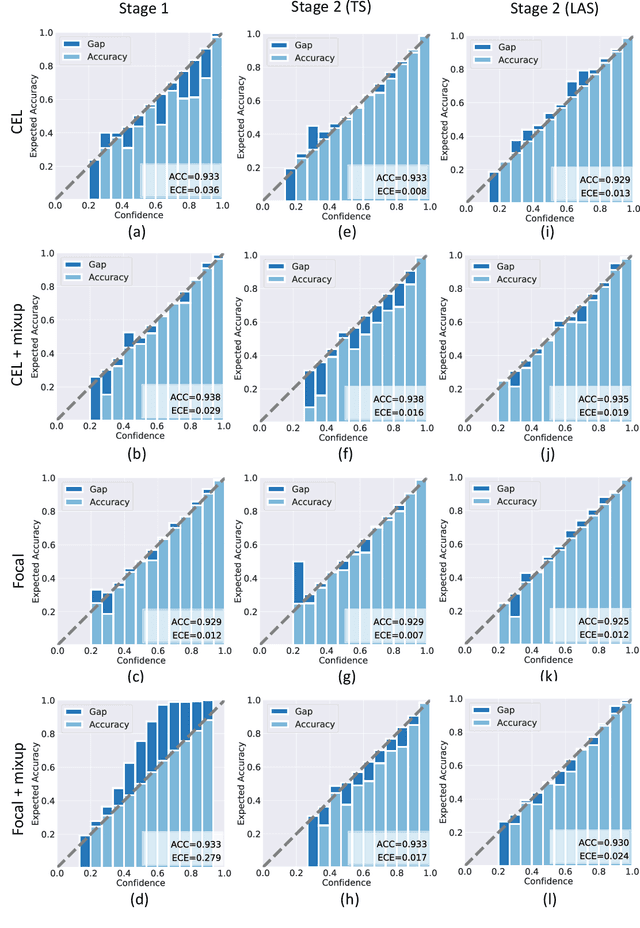
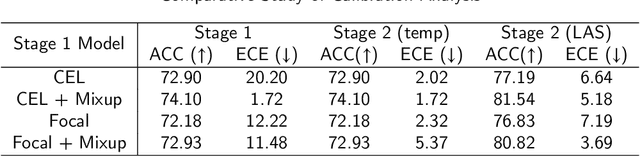
Abstract:Although deep learning prediction models have been successful in the discrimination of different classes, they can often suffer from poor calibration across challenging domains including healthcare. Moreover, the long-tail distribution poses great challenges in deep learning classification problems including clinical disease prediction. There are approaches proposed recently to calibrate deep prediction in computer vision, but there are no studies found to demonstrate how the representative models work in different challenging contexts. In this paper, we bridge the confidence calibration from computer vision to medical imaging with a comparative study of four high-impact calibration models. Our studies are conducted in different contexts (natural image classification and lung cancer risk estimation) including in balanced vs. imbalanced training sets and in computer vision vs. medical imaging. Our results support key findings: (1) We achieve new conclusions which are not studied under different learning contexts, e.g., combining two calibration models that both mitigate the overconfident prediction can lead to under-confident prediction, and simpler calibration models from the computer vision domain tend to be more generalizable to medical imaging. (2) We highlight the gap between general computer vision tasks and medical imaging prediction, e.g., calibration methods ideal for general computer vision tasks may in fact damage the calibration of medical imaging prediction. (3) We also reinforce previous conclusions in natural image classification settings. We believe that this study has merits to guide readers to choose calibration models and understand gaps between general computer vision and medical imaging domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge