Sanja L. Antic
Body Composition Assessment with Limited Field-of-view Computed Tomography: A Semantic Image Extension Perspective
Jul 13, 2022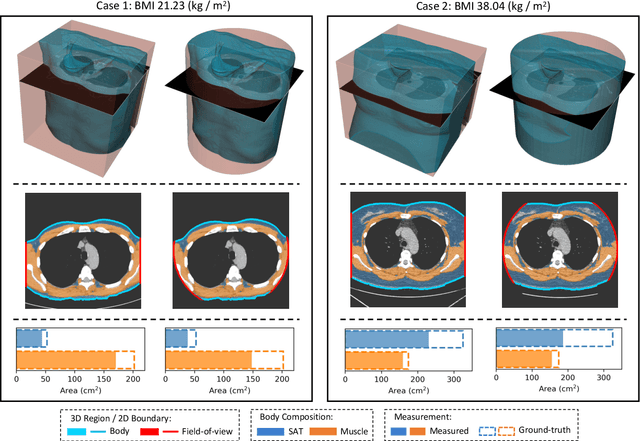
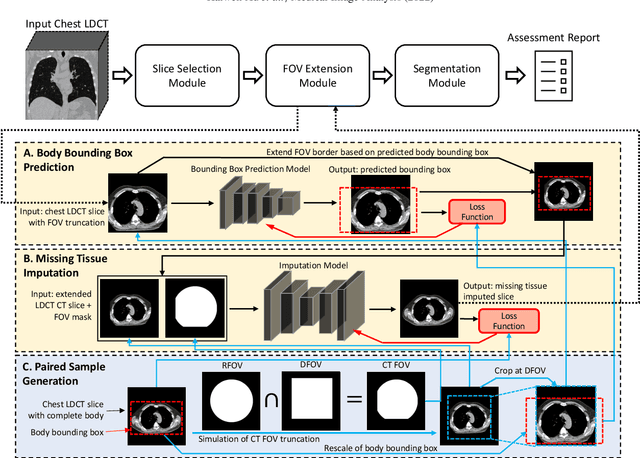
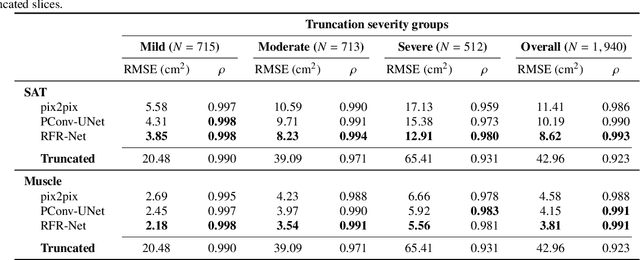
Abstract:Field-of-view (FOV) tissue truncation beyond the lungs is common in routine lung screening computed tomography (CT). This poses limitations for opportunistic CT- based body composition (BC) assessment as key anatomical structures are missing. Traditionally, extending the FOV of CT is considered as a CT reconstruction problem using limited data. However, this approach relies on the projection domain data which might not be available in application. In this work, we formulate the problem from the semantic image extension perspective which only requires image data as inputs. The proposed two-stage method identifies a new FOV border based on the estimated extent of the complete body and imputes missing tissues in the truncated region. The training samples are simulated using CT slices with complete body in FOV, making the model development self-supervised. We evaluate the validity of the proposed method in automatic BC assessment using lung screening CT with limited FOV. The proposed method effectively restores the missing tissues and reduces BC assessment error introduced by FOV tissue truncation. In the BC assessment for a large-scale lung screening CT dataset, this correction improves both the intra-subject consistency and the correlation with anthropometric approximations. The developed method is available at https://github.com/MASILab/S-EFOV.
A Comparative Study of Confidence Calibration in Deep Learning: From Computer Vision to Medical Imaging
Jun 17, 2022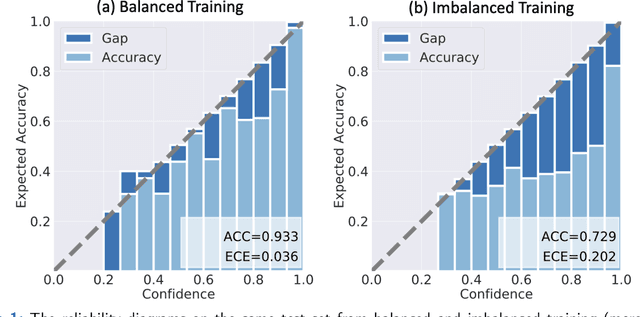
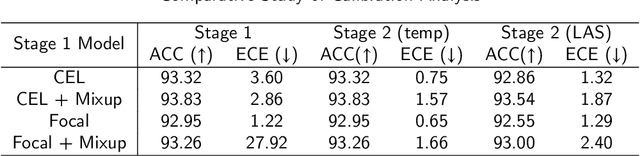
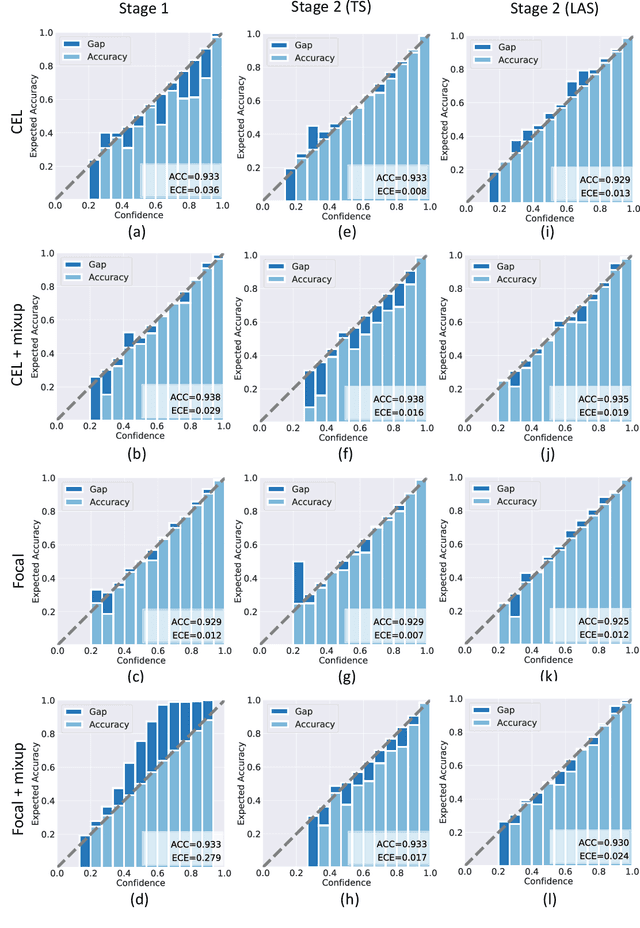
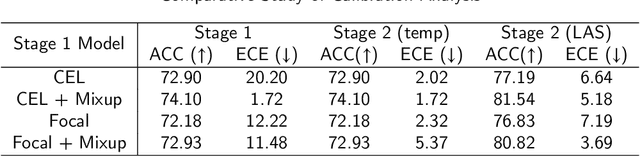
Abstract:Although deep learning prediction models have been successful in the discrimination of different classes, they can often suffer from poor calibration across challenging domains including healthcare. Moreover, the long-tail distribution poses great challenges in deep learning classification problems including clinical disease prediction. There are approaches proposed recently to calibrate deep prediction in computer vision, but there are no studies found to demonstrate how the representative models work in different challenging contexts. In this paper, we bridge the confidence calibration from computer vision to medical imaging with a comparative study of four high-impact calibration models. Our studies are conducted in different contexts (natural image classification and lung cancer risk estimation) including in balanced vs. imbalanced training sets and in computer vision vs. medical imaging. Our results support key findings: (1) We achieve new conclusions which are not studied under different learning contexts, e.g., combining two calibration models that both mitigate the overconfident prediction can lead to under-confident prediction, and simpler calibration models from the computer vision domain tend to be more generalizable to medical imaging. (2) We highlight the gap between general computer vision tasks and medical imaging prediction, e.g., calibration methods ideal for general computer vision tasks may in fact damage the calibration of medical imaging prediction. (3) We also reinforce previous conclusions in natural image classification settings. We believe that this study has merits to guide readers to choose calibration models and understand gaps between general computer vision and medical imaging domains.
Deep Multi-path Network Integrating Incomplete Biomarker and Chest CT Data for Evaluating Lung Cancer Risk
Oct 19, 2020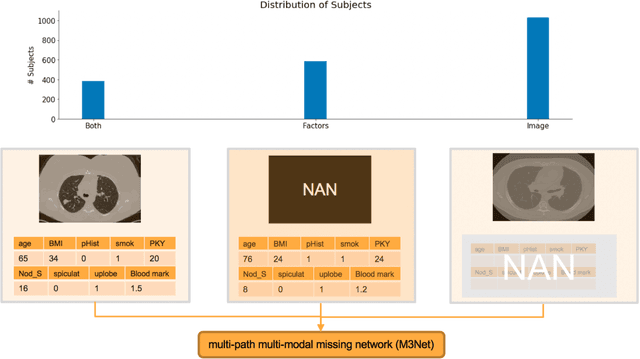
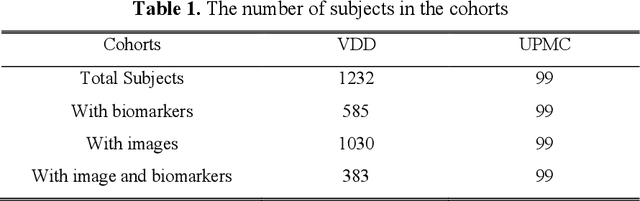
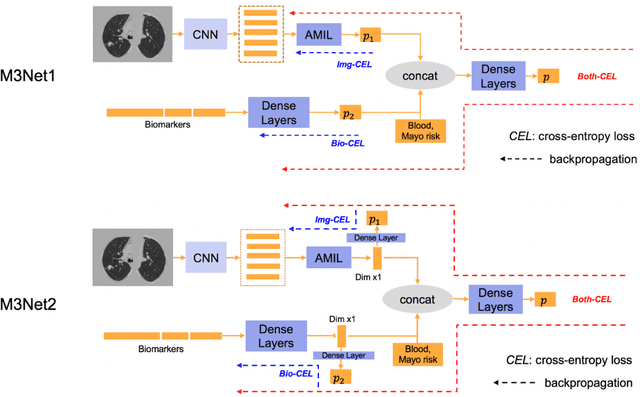
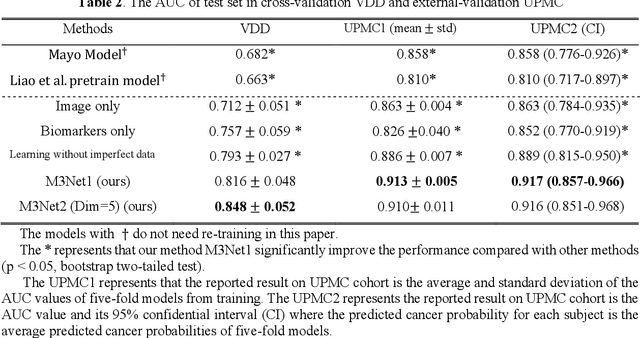
Abstract:Clinical data elements (CDEs) (e.g., age, smoking history), blood markers and chest computed tomography (CT) structural features have been regarded as effective means for assessing lung cancer risk. These independent variables can provide complementary information and we hypothesize that combining them will improve the prediction accuracy. In practice, not all patients have all these variables available. In this paper, we propose a new network design, termed as multi-path multi-modal missing network (M3Net), to integrate the multi-modal data (i.e., CDEs, biomarker and CT image) considering missing modality with multiple paths neural network. Each path learns discriminative features of one modality, and different modalities are fused in a second stage for an integrated prediction. The network can be trained end-to-end with both medical image features and CDEs/biomarkers, or make a prediction with single modality. We evaluate M3Net with datasets including three sites from the Consortium for Molecular and Cellular Characterization of Screen-Detected Lesions (MCL) project. Our method is cross validated within a cohort of 1291 subjects (383 subjects with complete CDEs/biomarkers and CT images), and externally validated with a cohort of 99 subjects (99 with complete CDEs/biomarkers and CT images). Both cross-validation and external-validation results show that combining multiple modality significantly improves the predicting performance of single modality. The results suggest that integrating subjects with missing either CDEs/biomarker or CT imaging features can contribute to the discriminatory power of our model (p < 0.05, bootstrap two-tailed test). In summary, the proposed M3Net framework provides an effective way to integrate image and non-image data in the context of missing information.
Internal-transfer Weighting of Multi-task Learning for Lung Cancer Detection
Dec 16, 2019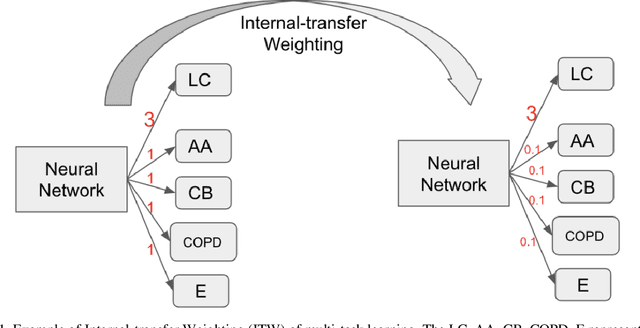
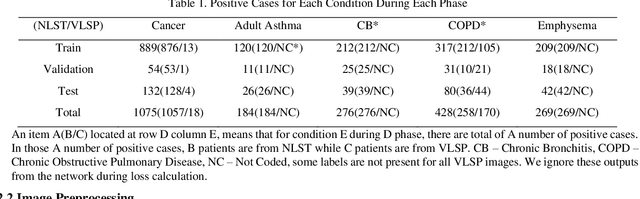
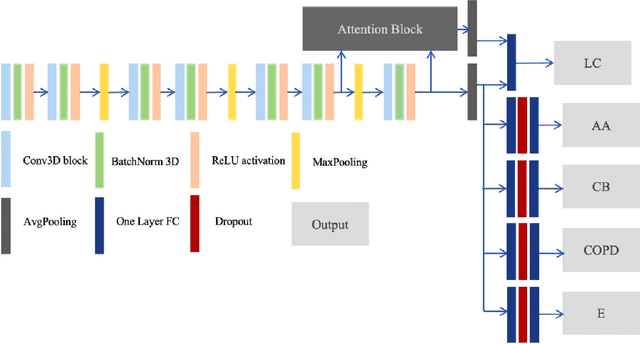
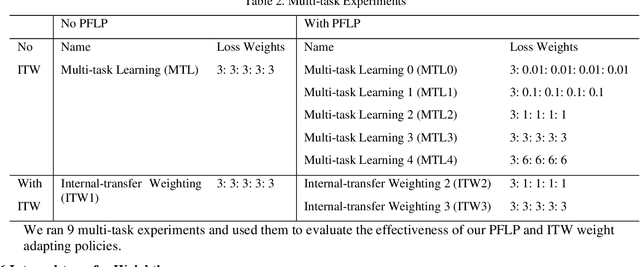
Abstract:Recently, multi-task networks have shown to both offer additional estimation capabilities, and, perhaps more importantly, increased performance over single-task networks on a "main/primary" task. However, balancing the optimization criteria of multi-task networks across different tasks is an area of active exploration. Here, we extend a previously proposed 3D attention-based network with four additional multi-task subnetworks for the detection of lung cancer and four auxiliary tasks (diagnosis of asthma, chronic bronchitis, chronic obstructive pulmonary disease, and emphysema). We introduce and evaluate a learning policy, Periodic Focusing Learning Policy (PFLP), that alternates the dominance of tasks throughout the training. To improve performance on the primary task, we propose an Internal-Transfer Weighting (ITW) strategy to suppress the loss functions on auxiliary tasks for the final stages of training. To evaluate this approach, we examined 3386 patients (single scan per patient) from the National Lung Screening Trial (NLST) and de-identified data from the Vanderbilt Lung Screening Program, with a 2517/277/592 (scans) split for training, validation, and testing. Baseline networks include a single-task strategy and a multi-task strategy without adaptive weights (PFLP/ITW), while primary experiments are multi-task trials with either PFLP or ITW or both. On the test set for lung cancer prediction, the baseline single-task network achieved prediction AUC of 0.8080 and the multi-task baseline failed to converge (AUC 0.6720). However, applying PFLP helped multi-task network clarify and achieved test set lung cancer prediction AUC of 0.8402. Furthermore, our ITW technique boosted the PFLP enabled multi-task network and achieved an AUC of 0.8462 (McNemar test, p < 0.01).
Deep Multi-task Prediction of Lung Cancer and Cancer-free Progression from Censored Heterogenous Clinical Imaging
Nov 12, 2019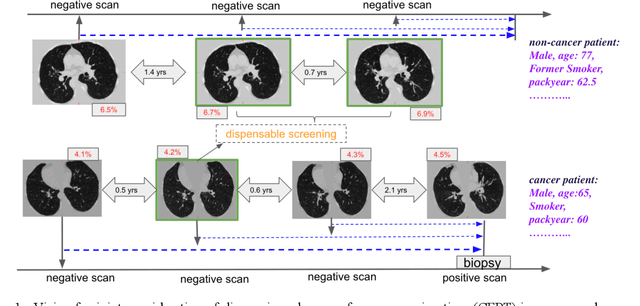
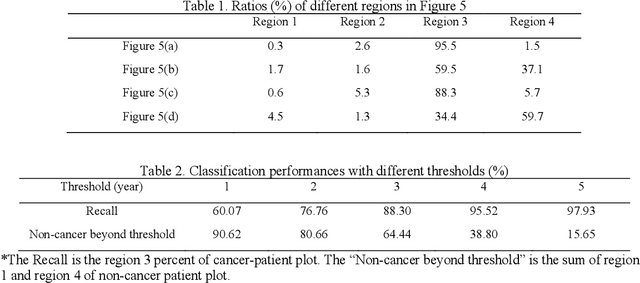
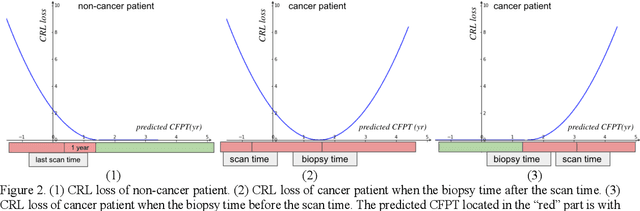
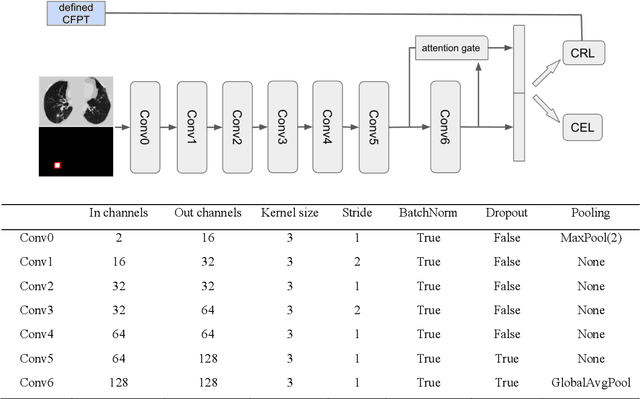
Abstract:Annual low dose computed tomography (CT) lung screening is currently advised for individuals at high risk of lung cancer (e.g., heavy smokers between 55 and 80 years old). The recommended screening practice significantly reduces all-cause mortality, but the vast majority of screening results are negative for cancer. If patients at very low risk could be identified based on individualized, image-based biomarkers, the health care resources could be more efficiently allocated to higher risk patients and reduce overall exposure to ionizing radiation. In this work, we propose a multi-task (diagnosis and prognosis) deep convolutional neural network to improve the diagnostic accuracy over a baseline model while simultaneously estimating a personalized cancer-free progression time (CFPT). A novel Censored Regression Loss (CRL) is proposed to perform weakly supervised regression so that even single negative screening scans can provide small incremental value. Herein, we study 2287 scans from 1433 de-identified patients from the Vanderbilt Lung Screening Program (VLSP) and Molecular Characterization Laboratories (MCL) cohorts. Using five-fold cross-validation, we train a 3D attention-based network under two scenarios: (1) single-task learning with only classification, and (2) multi-task learning with both classification and regression. The single-task learning leads to a higher AUC compared with the Kaggle challenge winner pre-trained model (0.878 v. 0.856), and multi-task learning significantly improves the single-task one (AUC 0.895, p<0.01, McNemar test). In summary, the image-based predicted CFPT can be used in follow-up year lung cancer prediction and data assessment.
Distanced LSTM: Time-Distanced Gates in Long Short-Term Memory Models for Lung Cancer Detection
Sep 11, 2019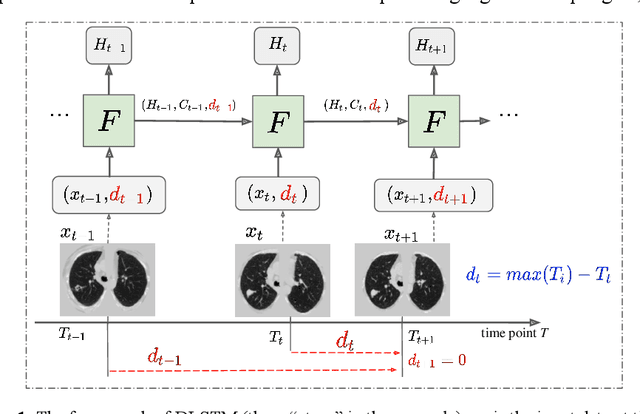
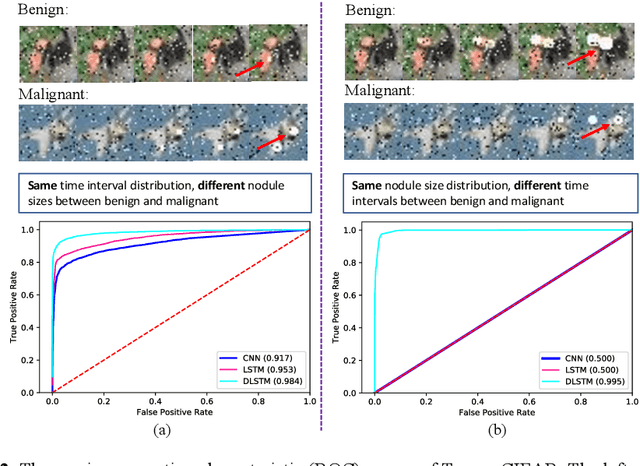
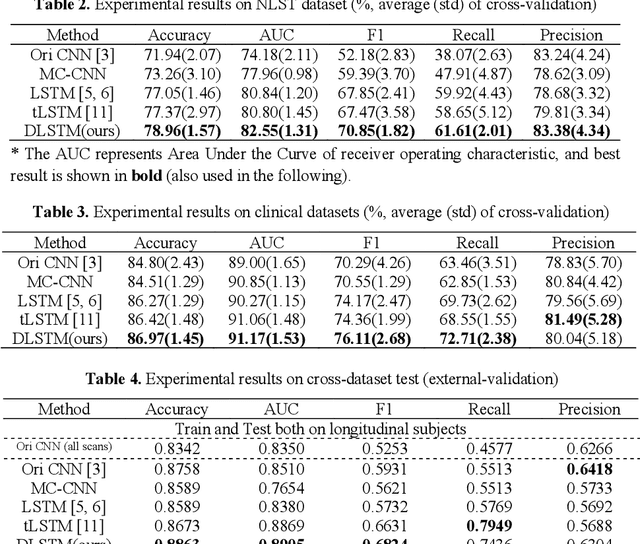
Abstract:The field of lung nodule detection and cancer prediction has been rapidly developing with the support of large public data archives. Previous studies have largely focused on cross-sectional (single) CT data. Herein, we consider longitudinal data. The Long Short-Term Memory (LSTM) model addresses learning with regularly spaced time points (i.e., equal temporal intervals). However, clinical imaging follows patient needs with often heterogeneous, irregular acquisitions. To model both regular and irregular longitudinal samples, we generalize the LSTM model with the Distanced LSTM (DLSTM) for temporally varied acquisitions. The DLSTM includes a Temporal Emphasis Model (TEM) that enables learning across regularly and irregularly sampled intervals. Briefly, (1) the time intervals between longitudinal scans are modeled explicitly, (2) temporally adjustable forget and input gates are introduced for irregular temporal sampling; and (3) the latest longitudinal scan has an additional emphasis term. We evaluate the DLSTM framework in three datasets including simulated data, 1794 National Lung Screening Trial (NLST) scans, and 1420 clinically acquired data with heterogeneous and irregular temporal accession. The experiments on the first two datasets demonstrate that our method achieves competitive performance on both simulated and regularly sampled datasets (e.g. improve LSTM from 0.6785 to 0.7085 on F1 score in NLST). In external validation of clinically and irregularly acquired data, the benchmarks achieved 0.8350 (CNN feature) and 0.8380 (LSTM) on the area under the ROC curve (AUC) score, while the proposed DLSTM achieves 0.8905.
Lung Cancer Detection using Co-learning from Chest CT Images and Clinical Demographics
Feb 21, 2019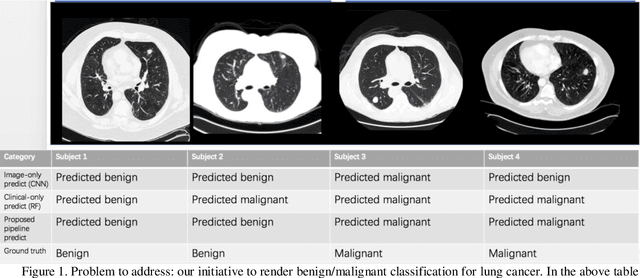
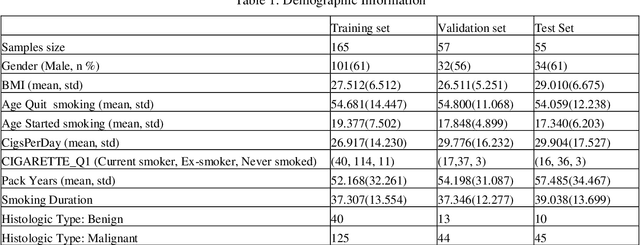
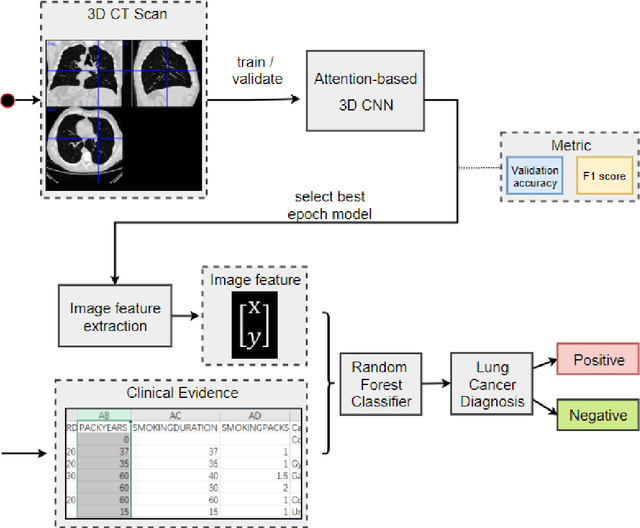
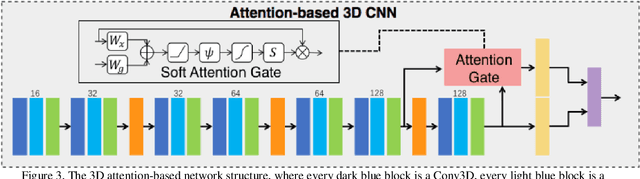
Abstract:Early detection of lung cancer is essential in reducing mortality. Recent studies have demonstrated the clinical utility of low-dose computed tomography (CT) to detect lung cancer among individuals selected based on very limited clinical information. However, this strategy yields high false positive rates, which can lead to unnecessary and potentially harmful procedures. To address such challenges, we established a pipeline that co-learns from detailed clinical demographics and 3D CT images. Toward this end, we leveraged data from the Consortium for Molecular and Cellular Characterization of Screen-Detected Lesions (MCL), which focuses on early detection of lung cancer. A 3D attention-based deep convolutional neural net (DCNN) is proposed to identify lung cancer from the chest CT scan without prior anatomical location of the suspicious nodule. To improve upon the non-invasive discrimination between benign and malignant, we applied a random forest classifier to a dataset integrating clinical information to imaging data. The results show that the AUC obtained from clinical demographics alone was 0.635 while the attention network alone reached an accuracy of 0.687. In contrast when applying our proposed pipeline integrating clinical and imaging variables, we reached an AUC of 0.787 on the testing dataset. The proposed network both efficiently captures anatomical information for classification and also generates attention maps that explain the features that drive performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge