Thomas Z. Li
AdaFuse: Adaptive Multimodal Fusion for Lung Cancer Risk Prediction via Reinforcement Learning
Jan 30, 2026Abstract:Multimodal fusion has emerged as a promising paradigm for disease diagnosis and prognosis, integrating complementary information from heterogeneous data sources such as medical images, clinical records, and radiology reports. However, existing fusion methods process all available modalities through the network, either treating them equally or learning to assign different contribution weights, leaving a fundamental question unaddressed: for a given patient, should certain modalities be used at all? We present AdaFuse, an adaptive multimodal fusion framework that leverages reinforcement learning (RL) to learn patient-specific modality selection and fusion strategies for lung cancer risk prediction. AdaFuse formulates multimodal fusion as a sequential decision process, where the policy network iteratively decides whether to incorporate an additional modality or proceed to prediction based on the information already acquired. This sequential formulation enables the model to condition each selection on previously observed modalities and terminate early when sufficient information is available, rather than committing to a fixed subset upfront. We evaluate AdaFuse on the National Lung Screening Trial (NLST) dataset. Experimental results demonstrate that AdaFuse achieves the highest AUC (0.762) compared to the best single-modality baseline (0.732), the best fixed fusion strategy (0.759), and adaptive baselines including DynMM (0.754) and MoE (0.742), while using fewer FLOPs than all triple-modality methods. Our work demonstrates the potential of reinforcement learning for personalized multimodal fusion in medical imaging, representing a shift from uniform fusion strategies toward adaptive diagnostic pipelines that learn when to consult additional modalities and when existing information suffices for accurate prediction.
Self-supervised learning of imaging and clinical signatures using a multimodal joint-embedding predictive architecture
Sep 18, 2025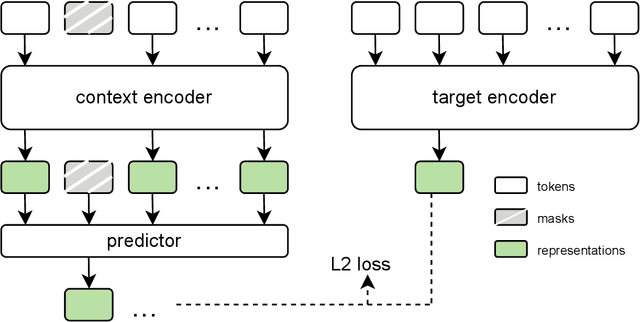

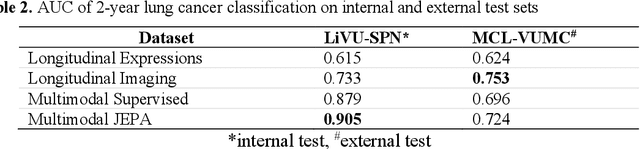
Abstract:The development of multimodal models for pulmonary nodule diagnosis is limited by the scarcity of labeled data and the tendency for these models to overfit on the training distribution. In this work, we leverage self-supervised learning from longitudinal and multimodal archives to address these challenges. We curate an unlabeled set of patients with CT scans and linked electronic health records from our home institution to power joint embedding predictive architecture (JEPA) pretraining. After supervised finetuning, we show that our approach outperforms an unregularized multimodal model and imaging-only model in an internal cohort (ours: 0.91, multimodal: 0.88, imaging-only: 0.73 AUC), but underperforms in an external cohort (ours: 0.72, imaging-only: 0.75 AUC). We develop a synthetic environment that characterizes the context in which JEPA may underperform. This work innovates an approach that leverages unlabeled multimodal medical archives to improve predictive models and demonstrates its advantages and limitations in pulmonary nodule diagnosis.
Cohort-Aware Agents for Individualized Lung Cancer Risk Prediction Using a Retrieval-Augmented Model Selection Framework
Aug 20, 2025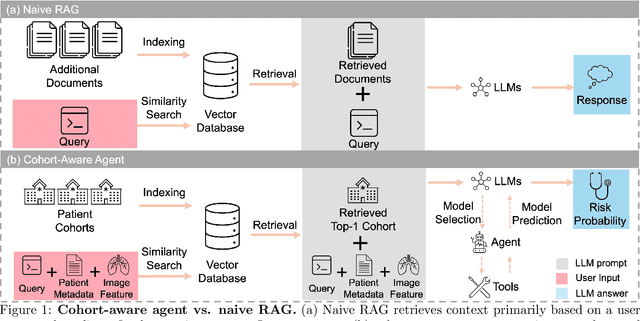
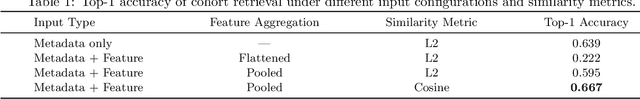
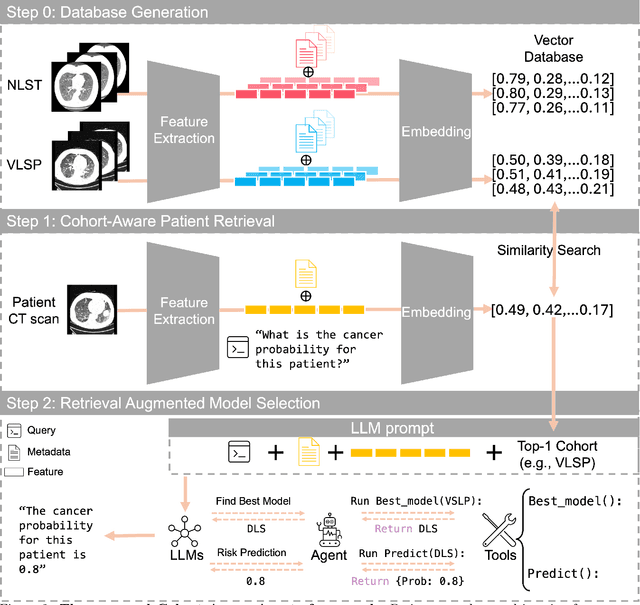
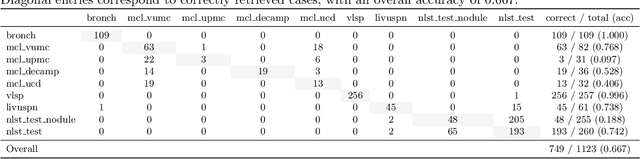
Abstract:Accurate lung cancer risk prediction remains challenging due to substantial variability across patient populations and clinical settings -- no single model performs best for all cohorts. To address this, we propose a personalized lung cancer risk prediction agent that dynamically selects the most appropriate model for each patient by combining cohort-specific knowledge with modern retrieval and reasoning techniques. Given a patient's CT scan and structured metadata -- including demographic, clinical, and nodule-level features -- the agent first performs cohort retrieval using FAISS-based similarity search across nine diverse real-world cohorts to identify the most relevant patient population from a multi-institutional database. Second, a Large Language Model (LLM) is prompted with the retrieved cohort and its associated performance metrics to recommend the optimal prediction algorithm from a pool of eight representative models, including classical linear risk models (e.g., Mayo, Brock), temporally-aware models (e.g., TDVIT, DLSTM), and multi-modal computer vision-based approaches (e.g., Liao, Sybil, DLS, DLI). This two-stage agent pipeline -- retrieval via FAISS and reasoning via LLM -- enables dynamic, cohort-aware risk prediction personalized to each patient's profile. Building on this architecture, the agent supports flexible and cohort-driven model selection across diverse clinical populations, offering a practical path toward individualized risk assessment in real-world lung cancer screening.
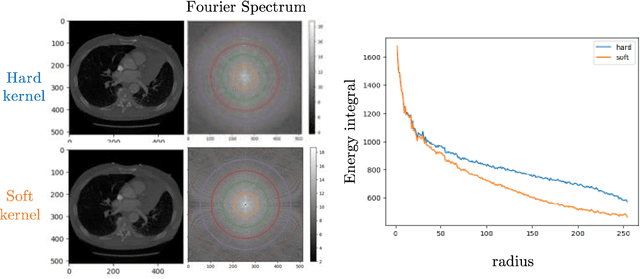
Multipath cycleGAN for harmonization of paired and unpaired low-dose lung computed tomography reconstruction kernels
May 28, 2025Abstract:Reconstruction kernels in computed tomography (CT) affect spatial resolution and noise characteristics, introducing systematic variability in quantitative imaging measurements such as emphysema quantification. Choosing an appropriate kernel is therefore essential for consistent quantitative analysis. We propose a multipath cycleGAN model for CT kernel harmonization, trained on a mixture of paired and unpaired data from a low-dose lung cancer screening cohort. The model features domain-specific encoders and decoders with a shared latent space and uses discriminators tailored for each domain.We train the model on 42 kernel combinations using 100 scans each from seven representative kernels in the National Lung Screening Trial (NLST) dataset. To evaluate performance, 240 scans from each kernel are harmonized to a reference soft kernel, and emphysema is quantified before and after harmonization. A general linear model assesses the impact of age, sex, smoking status, and kernel on emphysema. We also evaluate harmonization from soft kernels to a reference hard kernel. To assess anatomical consistency, we compare segmentations of lung vessels, muscle, and subcutaneous adipose tissue generated by TotalSegmentator between harmonized and original images. Our model is benchmarked against traditional and switchable cycleGANs. For paired kernels, our approach reduces bias in emphysema scores, as seen in Bland-Altman plots (p<0.05). For unpaired kernels, harmonization eliminates confounding differences in emphysema (p>0.05). High Dice scores confirm preservation of muscle and fat anatomy, while lung vessel overlap remains reasonable. Overall, our shared latent space multipath cycleGAN enables robust harmonization across paired and unpaired CT kernels, improving emphysema quantification and preserving anatomical fidelity.
Unsupervised Discovery of Clinical Disease Signatures Using Probabilistic Independence
Feb 08, 2024Abstract:Insufficiently precise diagnosis of clinical disease is likely responsible for many treatment failures, even for common conditions and treatments. With a large enough dataset, it may be possible to use unsupervised machine learning to define clinical disease patterns more precisely. We present an approach to learning these patterns by using probabilistic independence to disentangle the imprint on the medical record of causal latent sources of disease. We inferred a broad set of 2000 clinical signatures of latent sources from 9195 variables in 269,099 Electronic Health Records. The learned signatures produced better discrimination than the original variables in a lung cancer prediction task unknown to the inference algorithm, predicting 3-year malignancy in patients with no history of cancer before a solitary lung nodule was discovered. More importantly, the signatures' greater explanatory power identified pre-nodule signatures of apparently undiagnosed cancer in many of those patients.
Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures From Routine EHRs for Pulmonary Nodule Classification
Apr 10, 2023



Abstract:The accuracy of predictive models for solitary pulmonary nodule (SPN) diagnosis can be greatly increased by incorporating repeat imaging and medical context, such as electronic health records (EHRs). However, clinically routine modalities such as imaging and diagnostic codes can be asynchronous and irregularly sampled over different time scales which are obstacles to longitudinal multimodal learning. In this work, we propose a transformer-based multimodal strategy to integrate repeat imaging with longitudinal clinical signatures from routinely collected EHRs for SPN classification. We perform unsupervised disentanglement of latent clinical signatures and leverage time-distance scaled self-attention to jointly learn from clinical signatures expressions and chest computed tomography (CT) scans. Our classifier is pretrained on 2,668 scans from a public dataset and 1,149 subjects with longitudinal chest CTs, billing codes, medications, and laboratory tests from EHRs of our home institution. Evaluation on 227 subjects with challenging SPNs revealed a significant AUC improvement over a longitudinal multimodal baseline (0.824 vs 0.752 AUC), as well as improvements over a single cross-section multimodal scenario (0.809 AUC) and a longitudinal imaging-only scenario (0.741 AUC). This work demonstrates significant advantages with a novel approach for co-learning longitudinal imaging and non-imaging phenotypes with transformers.
Zero-shot CT Field-of-view Completion with Unconditional Generative Diffusion Prior
Apr 07, 2023Abstract:Anatomically consistent field-of-view (FOV) completion to recover truncated body sections has important applications in quantitative analyses of computed tomography (CT) with limited FOV. Existing solution based on conditional generative models relies on the fidelity of synthetic truncation patterns at training phase, which poses limitations for the generalizability of the method to potential unknown types of truncation. In this study, we evaluate a zero-shot method based on a pretrained unconditional generative diffusion prior, where truncation pattern with arbitrary forms can be specified at inference phase. In evaluation on simulated chest CT slices with synthetic FOV truncation, the method is capable of recovering anatomically consistent body sections and subcutaneous adipose tissue measurement error caused by FOV truncation. However, the correction accuracy is inferior to the conditionally trained counterpart.
Time-distance vision transformers in lung cancer diagnosis from longitudinal computed tomography
Sep 04, 2022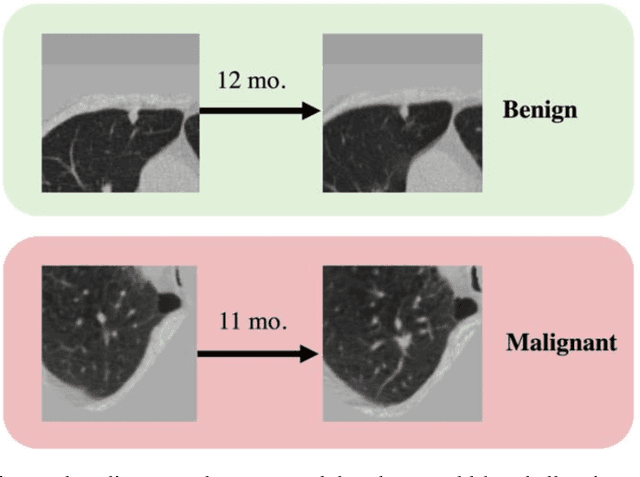

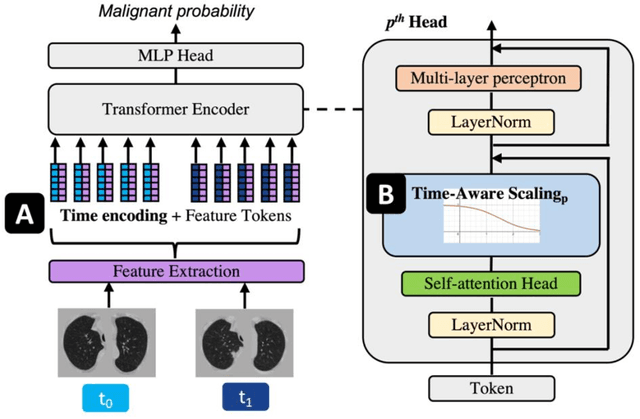

Abstract:Features learned from single radiologic images are unable to provide information about whether and how much a lesion may be changing over time. Time-dependent features computed from repeated images can capture those changes and help identify malignant lesions by their temporal behavior. However, longitudinal medical imaging presents the unique challenge of sparse, irregular time intervals in data acquisition. While self-attention has been shown to be a versatile and efficient learning mechanism for time series and natural images, its potential for interpreting temporal distance between sparse, irregularly sampled spatial features has not been explored. In this work, we propose two interpretations of a time-distance vision transformer (ViT) by using (1) vector embeddings of continuous time and (2) a temporal emphasis model to scale self-attention weights. The two algorithms are evaluated based on benign versus malignant lung cancer discrimination of synthetic pulmonary nodules and lung screening computed tomography studies from the National Lung Screening Trial (NLST). Experiments evaluating the time-distance ViTs on synthetic nodules show a fundamental improvement in classifying irregularly sampled longitudinal images when compared to standard ViTs. In cross-validation on screening chest CTs from the NLST, our methods (0.785 and 0.786 AUC respectively) significantly outperform a cross-sectional approach (0.734 AUC) and match the discriminative performance of the leading longitudinal medical imaging algorithm (0.779 AUC) on benign versus malignant classification. This work represents the first self-attention-based framework for classifying longitudinal medical images. Our code is available at https://github.com/tom1193/time-distance-transformer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge