Sokratis Makrogiannis
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Single Slice Thigh CT Muscle Group Segmentation with Domain Adaptation and Self-Training
Nov 30, 2022Abstract:Objective: Thigh muscle group segmentation is important for assessment of muscle anatomy, metabolic disease and aging. Many efforts have been put into quantifying muscle tissues with magnetic resonance (MR) imaging including manual annotation of individual muscles. However, leveraging publicly available annotations in MR images to achieve muscle group segmentation on single slice computed tomography (CT) thigh images is challenging. Method: We propose an unsupervised domain adaptation pipeline with self-training to transfer labels from 3D MR to single CT slice. First, we transform the image appearance from MR to CT with CycleGAN and feed the synthesized CT images to a segmenter simultaneously. Single CT slices are divided into hard and easy cohorts based on the entropy of pseudo labels inferenced by the segmenter. After refining easy cohort pseudo labels based on anatomical assumption, self-training with easy and hard splits is applied to fine tune the segmenter. Results: On 152 withheld single CT thigh images, the proposed pipeline achieved a mean Dice of 0.888(0.041) across all muscle groups including sartorius, hamstrings, quadriceps femoris and gracilis. muscles Conclusion: To our best knowledge, this is the first pipeline to achieve thigh imaging domain adaptation from MR to CT. The proposed pipeline is effective and robust in extracting muscle groups on 2D single slice CT thigh images.The container is available for public use at https://github.com/MASILab/DA_CT_muscle_seg
AI in Osteoporosis
Sep 22, 2021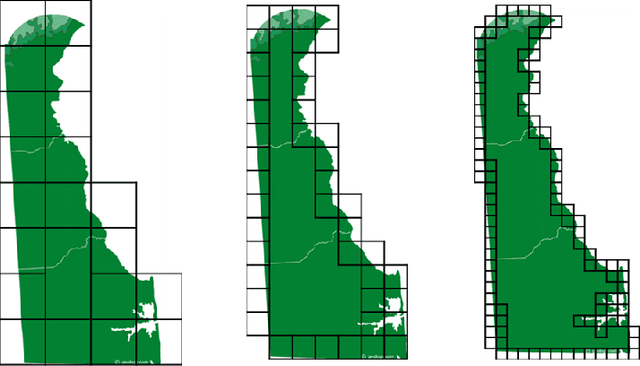
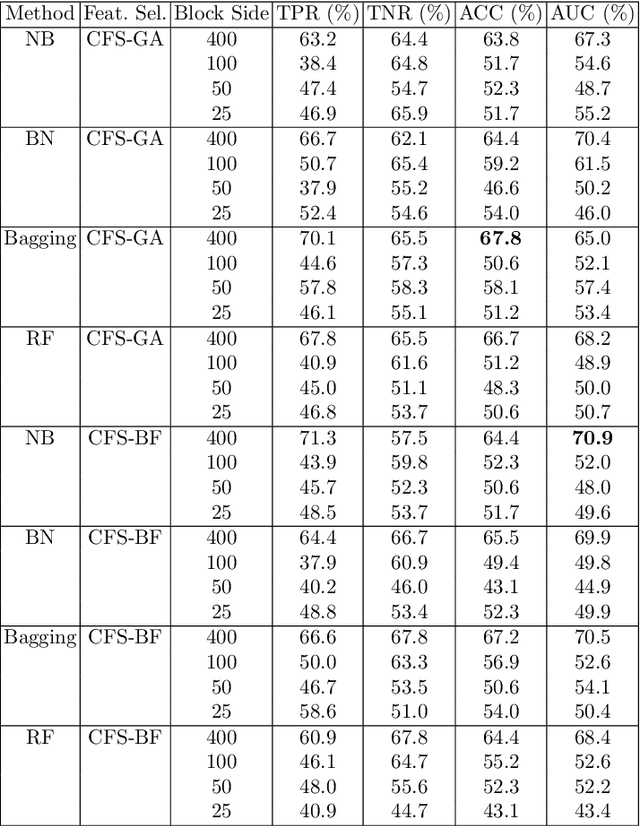

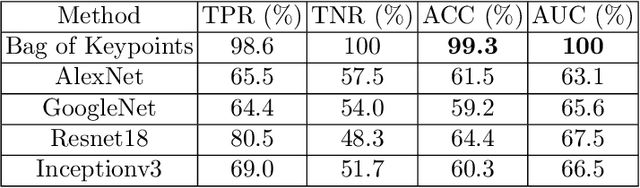
Abstract:In this chapter we explore and evaluate methods for trabecular bone characterization and osteoporosis diagnosis with increased interest in sparse approximations. We first describe texture representation and classification techniques, patch-based methods such as Bag of Keypoints, and more recent deep neural networks. Then we introduce the concept of sparse representations for pattern recognition and we detail integrative sparse analysis methods and classifier decision fusion methods. We report cross-validation results on osteoporosis datasets of bone radiographs and compare the results produced by the different categories of methods. We conclude that advances in the AI and machine learning fields have enabled the development of methods that can be used as diagnostic tools in clinical settings.
Discriminative Localized Sparse Representations for Breast Cancer Screening
Nov 20, 2020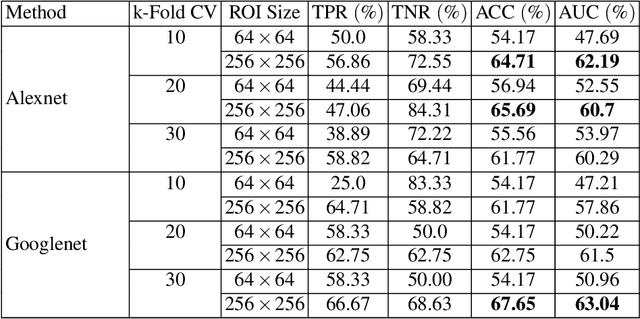
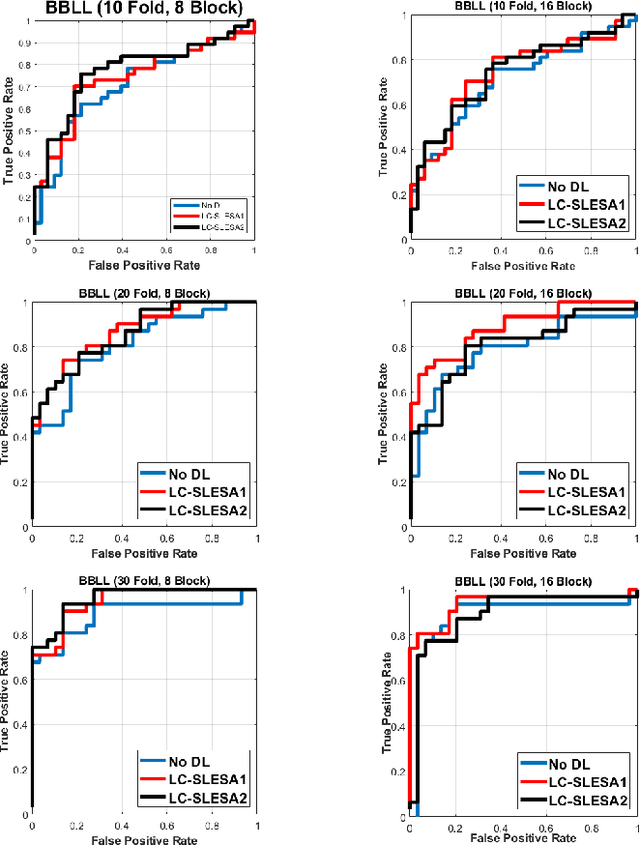
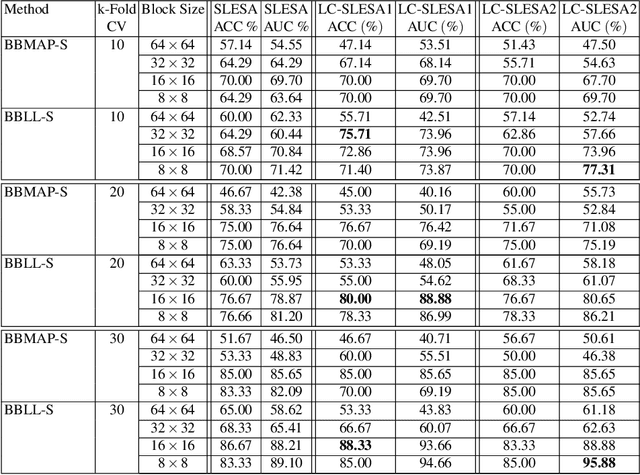

Abstract:Breast cancer is the most common cancer among women both in developed and developing countries. Early detection and diagnosis of breast cancer may reduce its mortality and improve the quality of life. Computer-aided detection (CADx) and computer-aided diagnosis (CAD) techniques have shown promise for reducing the burden of human expert reading and improve the accuracy and reproducibility of results. Sparse analysis techniques have produced relevant results for representing and recognizing imaging patterns. In this work we propose a method for Label Consistent Spatially Localized Ensemble Sparse Analysis (LC-SLESA). In this work we apply dictionary learning to our block based sparse analysis method to classify breast lesions as benign or malignant. The performance of our method in conjunction with LC-KSVD dictionary learning is evaluated using 10-, 20-, and 30-fold cross validation on the MIAS dataset. Our results indicate that the proposed sparse analyses may be a useful component for breast cancer screening applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge